Proteomic Profiling of Small Extracellular Vesicles Secreted by Human Pancreatic Cancer Cells Implicated in Cellular Transformation
- PMID: 32382024
- PMCID: PMC7205864
- DOI: 10.1038/s41598-020-64718-6
Proteomic Profiling of Small Extracellular Vesicles Secreted by Human Pancreatic Cancer Cells Implicated in Cellular Transformation
Abstract
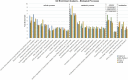
Extracellular vesicles secreted from tumor cells are functional vehicles capable of contributing to intercellular communication and metastasis. A growing number of studies have focused on elucidating the role that tumor-derived extracellular vesicles play in spreading pancreatic cancer to other organs, due to the highly metastatic nature of the disease. We recently showed that small extracellular vesicles secreted from pancreatic cancer cells could initiate malignant transformation of healthy cells. Here, we analyzed the protein cargo contained within these vesicles using mass spectrometry-based proteomics to better understand their makeup and biological characteristics. Three different human pancreatic cancer cell lines were compared to normal pancreatic epithelial cells revealing distinct differences in protein cargo between cancer and normal vesicles. Vesicles from cancer cells contain an enrichment of proteins that function in the endosomal compartment of cells responsible for vesicle formation and secretion in addition to proteins that have been shown to contribute to oncogenic cell transformation. Conversely, vesicles from normal pancreatic cells were shown to be enriched for immune response proteins. Collectively, results contribute to what we know about the cargo contained within or excluded from cancer cell-derived extracellular vesicles, supporting their role in biological processes including metastasis and cancer progression.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The Role of Extracellular Vesicles: An Epigenetic View of the Cancer Microenvironment.Biomed Res Int. 2015;2015:649161. doi: 10.1155/2015/649161. Epub 2015 Oct 25. Biomed Res Int. 2015. PMID: 26582468 Free PMC article. Review.
-
Oncogenic and Non-Malignant Pancreatic Exosome Cargo Reveal Distinct Expression of Oncogenic and Prognostic Factors Involved in Tumor Invasion and Metastasis.Proteomics. 2019 Apr;19(8):e1800158. doi: 10.1002/pmic.201800158. Epub 2019 Apr 9. Proteomics. 2019. PMID: 30893511
-
Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo.Sci Rep. 2016 Sep 8;6:32643. doi: 10.1038/srep32643. Sci Rep. 2016. PMID: 27605433 Free PMC article.
-
Proteomes of Extracellular Vesicles From Pancreatic Cancer Cells and Cancer-Associated Fibroblasts.Pancreas. 2022 Aug 1;51(7):790-799. doi: 10.1097/MPA.0000000000002115. Pancreas. 2022. PMID: 36395405 Free PMC article.
-
Exosomes in cancer development.Curr Opin Genet Dev. 2021 Feb;66:83-92. doi: 10.1016/j.gde.2020.12.018. Epub 2021 Jan 18. Curr Opin Genet Dev. 2021. PMID: 33477017 Review.
Cited by
-
Application of Mass Spectrometry in Pancreatic Cancer Translational Research.Front Oncol. 2021 Oct 11;11:667427. doi: 10.3389/fonc.2021.667427. eCollection 2021. Front Oncol. 2021. PMID: 34707986 Free PMC article. Review.
-
Proteomics in pancreatic cancer.Biomark Res. 2025 Jul 6;13(1):93. doi: 10.1186/s40364-025-00805-y. Biomark Res. 2025. PMID: 40619414 Free PMC article. Review.
-
A comparative Proteomics Analysis Identified Differentially Expressed Proteins in Pancreatic Cancer-Associated Stellate Cell Small Extracellular Vesicles.Mol Cell Proteomics. 2022 Dec;21(12):100438. doi: 10.1016/j.mcpro.2022.100438. Epub 2022 Nov 2. Mol Cell Proteomics. 2022. PMID: 36332889 Free PMC article.
-
Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review.Signal Transduct Target Ther. 2023 Mar 24;8(1):139. doi: 10.1038/s41392-023-01376-w. Signal Transduct Target Ther. 2023. PMID: 36964133 Free PMC article.
-
Atrioventricular re-entrant tachycardia and atrioventricular node re-entrant tachycardia in a patient with cancer under chemotherapy: a case report and literature review.Front Cardiovasc Med. 2024 Jun 7;11:1367893. doi: 10.3389/fcvm.2024.1367893. eCollection 2024. Front Cardiovasc Med. 2024. PMID: 38911514 Free PMC article.
References
-
- Thery C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

