Identification of differentially expressed genes and biological pathways in para-carcinoma tissues of HCC with different metastatic potentials
- PMID: 32382332
- PMCID: PMC7202278
- DOI: 10.3892/ol.2020.11493
Identification of differentially expressed genes and biological pathways in para-carcinoma tissues of HCC with different metastatic potentials
Abstract
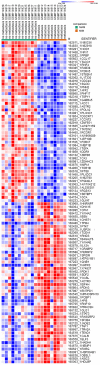
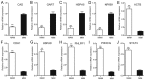
Hepatocellular carcinoma (HCC) is a malignant tumor with extensive metastasis. Changes in the tumor microenvironment provide favorable conditions for tumor metastasis. However, the role of changes to the tumor microenvironment in HCC metastasis is yet to be elucidated. The Gene Expression Omnibus expression profile GSE5093 consists of 20 noncancerous tissues surrounding HCC tissues, including 9 metastasis-inclined microenvironment samples with detectable metastases and 11 metastasis-averse microenvironment samples without detectable metastases. The present study assessed 35 HCC samples to verify the results of chip analysis. In total, 712 upregulated and 459 downregulated genes were identified, with 1,033 nodes, 7,589 edges and 10 hub genes. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed that the differentially expressed genes were significantly enriched in 'cell-cell adhesion', 'cell proliferation' and 'protein binding'. The top 10 hub genes were identified via a protein-protein interaction analysis. The 3 most significant modules were identified from the protein-protein network. Moreover, an association between hub genes and patient prognosis was identified. In conclusion, these candidate genes and pathways may help elucidate the mechanisms underlying HCC metastasis and identify more options for targeted therapy.
Keywords: differentially expressed genes; enrichment analysis; hepatocellular carcinoma; prognosis; protein-protein interaction; tumor microenvironment.
Copyright: © Liu et al.
Figures






References
LinkOut - more resources
Full Text Sources
