13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking
- PMID: 32384149
- PMCID: PMC7498303
- DOI: 10.1182/blood.2019004684
13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking
Abstract
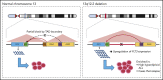
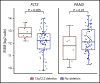
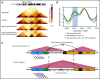
Mutations in the FMS-like tyrosine kinase 3 (FLT3) gene in 13q12.2 are among the most common driver events in acute leukemia, leading to increased cell proliferation and survival through activation of the phosphatidylinositol 3-kinase/AKT-, RAS/MAPK-, and STAT5-signaling pathways. In this study, we examine the pathogenetic impact of somatic hemizygous 13q12.2 microdeletions in B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) using 5 different patient cohorts (in total including 1418 cases). The 13q12.2 deletions occur immediately 5' of FLT3 and involve the PAN3 locus. By detailed analysis of the 13q12.2 segment, we show that the deletions lead to loss of a topologically associating domain border and an enhancer of FLT3. This results in increased cis interactions between the FLT3 promoter and another enhancer located distally to the deletion breakpoints, with subsequent allele-specific upregulation of FLT3 expression, expected to lead to ligand-independent activation of the receptor and downstream signaling. The 13q12.2 deletions are highly enriched in the high-hyperdiploid BCP ALL subtype (frequency 3.9% vs 0.5% in other BCP ALL) and in cases that subsequently relapsed. Taken together, our study describes a novel mechanism of FLT3 involvement in leukemogenesis by upregulation via chromatin remodeling and enhancer hijacking. These data further emphasize the role of FLT3 as a driver gene in BCP ALL.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


Comment in
-
Deletions in BCP-ALL result in a TAD more FLT3.Blood. 2020 Aug 20;136(8):919-920. doi: 10.1182/blood.2020006607. Blood. 2020. PMID: 32818246 Free PMC article.
-
13q12.2 deletions and FLT3 overexpression in acute leukemias.Blood Adv. 2021 Apr 27;5(8):2075-2078. doi: 10.1182/bloodadvances.2020003643. Blood Adv. 2021. PMID: 33877294 Free PMC article. No abstract available.
References
-
- Armstrong SA, Mabon ME, Silverman LB, et al. . FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103(9):3544-3546. - PubMed
-
- Paulsson K, Lilljebjörn H, Biloglav A, et al. . The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47(6):672-676. - PubMed
-
- Olsson L, Castor A, Behrendtz M, et al. . Deletions of IKZF1 and SPRED1 are associated with poor prognosis in a population-based series of pediatric B-cell precursor acute lymphoblastic leukemia diagnosed between 1992 and 2011. Leukemia. 2014;28(2):302-310. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

