Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors
- PMID: 32384610
- PMCID: PMC7288465
- DOI: 10.3390/genes11050511
Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors
Abstract
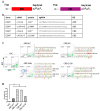
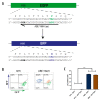
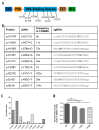
In contrast to CRISPR/Cas9 nucleases, CRISPR base editors (BE) and prime editors (PE) enable predefined nucleotide exchanges in genomic sequences without generating DNA double strand breaks. Here, we employed BE and PE mRNAs in conjunction with chemically synthesized sgRNAs and pegRNAs for efficient editing of human induced pluripotent stem cells (iPSC). Whereas we were unable to correct a disease-causing mutation in patient derived iPSCs using a CRISPR/Cas9 nuclease approach, we corrected the mutation back to wild type with high efficiency utilizing an adenine BE. We also used adenine and cytosine BEs to introduce nine different cancer associated TP53 mutations into human iPSCs with up to 90% efficiency, generating a panel of cell lines to investigate the biology of these mutations in an isogenic background. Finally, we pioneered the use of prime editing in human iPSCs, opening this important cell type for the precise modification of nucleotides not addressable by BEs and to multiple nucleotide exchanges. These approaches eliminate the necessity of deriving disease specific iPSCs from human donors and allows the comparison of different disease-causing mutations in isogenic genetic backgrounds.
Keywords: CRISPR/Cas9; base editors; human induced pluripotent stem cells; mRNA; prime editors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

