Integrative Cluster Analysis of Whole Hearts Reveals Proliferative Cardiomyocytes in Adult Mice
- PMID: 32384695
- PMCID: PMC7291011
- DOI: 10.3390/cells9051144
Integrative Cluster Analysis of Whole Hearts Reveals Proliferative Cardiomyocytes in Adult Mice
Abstract
The recent development and broad application of sequencing techniques at the single-cell level is generating an unprecedented amount of data. The different techniques have their individual limits, but the datasets also offer unexpected possibilities when utilized collectively. Here, we applied snRNA-seq in whole adult murine hearts from an inbred (C57BL/6NRj) and an outbred (Fzt:DU) mouse strain to directly compare the data with the publicly available scRNA-seq data of the tabula muris project. Explicitly choosing a single-nucleus approach allowed us to pin down the typical heart tissue-specific technical bias, coming up with novel insights on the mammalian heart cell composition. For our integrated dataset, cardiomyocytes, fibroblasts, and endothelial cells constituted the three main cell populations accounting for about 75% of all cells. However, their numbers severely differed between the individual datasets, with cardiomyocyte proportions ranging from about 9% in the tabula muris data to around 23% for our BL6 data, representing the prime example for cell capture technique related bias when using a conventional single-cell approach for these large cells. Most strikingly in our comparison was the discovery of a minor population of cardiomyocytes characterized by proliferation markers that could not be identified by analyzing the datasets individually. It is now widely accepted that the heart has an, albeit very restricted, regenerative potential. However there is still an ongoing debate where new cardiomyocytes arise from. Our findings support the idea that the renewal of the cardiomyocyte pool is driven by cytokinesis of resident cardiomyocytes rather than differentiation of progenitor cells. We thus provide data that can contribute to an understanding of heart cell regeneration, which is a prerequisite for future applications to enhance the process of heart repair.
Keywords: cytokinesis; harmony; heart regeneration; proliferation; single-cell RNA-Seq; single-nucleus RNA-Seq.
Conflict of interest statement
The authors declare no conflict of interest. The funders were not involved in study design, data collection and interpretation, and manuscript preparation.
Figures
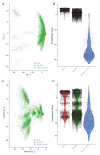
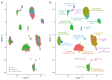



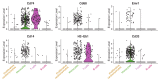
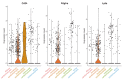

References
-
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

