Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice
- PMID: 32384927
- PMCID: PMC7206744
- DOI: 10.1186/s12870-020-02417-0
Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice
Abstract
Background: The interactions between Growth-regulating factors (GRFs) and GRF-Interacting Factors (GIFs) have been well demonstrated but it remains unclear whether different combinations of GRF and GIF play distinctive roles in the pathway downstream of the complex.
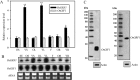
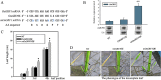
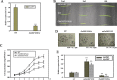
Results: Here we showed that OsGRF1 and OsGIF1 synergistically regulate leaf growth in rice. The expression of OsGIF1 emerged in all tissues with much higher level while that of OsGRF1 appeared preferentially only in the stem tips containing shoot apical meristem (SAM) and younger leaves containing leaf primordium. Overexpression of an OsmiR396-resistant version of mOsGRF1 resulted in expanded leaves due to increased cell proliferation while knockdown of OsGRF1 displayed an opposite phenotype. Overexpression of OsGIF1 did not exhibit new phenotype while knockdown lines displayed pleiotropic growth defects including shrunken leaves. The crossed lines of mOsGRF1 overexpression and OsGIF1 knockdown still exhibited shrunk leaves, indicating that OsGIF1 is indispensable in leaf growth regulated by OsGRF1. The expression of OsGRF1 could be upregulated by gibberellins (GAs) and downregulated by various stresses while that of OsGIF1 could not.
Conclusion: Our results suggest that OsGIF1 is in an excessive expression in various tissues and play roles in various aspects of growth while OsGRF1 may specifically involve in leaf growth through titrating OsGIF1. Both internal and external conditions impacting leaf growth are likely via way of regulating the expression of OsGRF1.
Keywords: Leaf growth; OsGIF1; OsGRF1; Stress response; miR396.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
OsGIF1 Positively Regulates the Sizes of Stems, Leaves, and Grains in Rice.Front Plant Sci. 2017 Oct 5;8:1730. doi: 10.3389/fpls.2017.01730. eCollection 2017. Front Plant Sci. 2017. PMID: 29051769 Free PMC article.
-
OsGRF1 and OsGRF2 play unequal redundant roles in regulating leaf vascular bundle formation.J Exp Bot. 2025 May 8:eraf193. doi: 10.1093/jxb/eraf193. Online ahead of print. J Exp Bot. 2025. PMID: 40338048
-
Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.).Plant Cell Physiol. 2004 Jul;45(7):897-904. doi: 10.1093/pcp/pch098. Plant Cell Physiol. 2004. PMID: 15295073
-
The Rice miR396-GRF-GIF-SWI/SNF Module: A Player in GA Signaling.Front Plant Sci. 2022 Jan 11;12:786641. doi: 10.3389/fpls.2021.786641. eCollection 2021. Front Plant Sci. 2022. PMID: 35087553 Free PMC article. Review.
-
MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology.Curr Opin Plant Biol. 2020 Feb;53:31-42. doi: 10.1016/j.pbi.2019.09.008. Epub 2019 Nov 11. Curr Opin Plant Biol. 2020. PMID: 31726426 Review.
Cited by
-
OsLC1, a transaldolase, regulates cell patterning and leaf morphology through modulation of secondary metabolism.Plant Biotechnol J. 2025 May;23(5):1751-1767. doi: 10.1111/pbi.70004. Epub 2025 Feb 14. Plant Biotechnol J. 2025. PMID: 39950420 Free PMC article.
-
Rice GROWTH-REGULATING FACTOR7 Modulates Plant Architecture through Regulating GA and Indole-3-Acetic Acid Metabolism.Plant Physiol. 2020 Sep;184(1):393-406. doi: 10.1104/pp.20.00302. Epub 2020 Jun 24. Plant Physiol. 2020. PMID: 32581114 Free PMC article.
-
Charged Gold Nanoparticles Promote In Vitro Proliferation in Nardostachys jatamansi by Differentially Regulating Chlorophyll Content, Hormone Concentration, and Antioxidant Activity.Antioxidants (Basel). 2022 Sep 30;11(10):1962. doi: 10.3390/antiox11101962. Antioxidants (Basel). 2022. PMID: 36290684 Free PMC article.
-
Analysis of Glucosinolate Content and Metabolism Related Genes in Different Parts of Chinese Flowering Cabbage.Front Plant Sci. 2022 Jan 17;12:767898. doi: 10.3389/fpls.2021.767898. eCollection 2021. Front Plant Sci. 2022. PMID: 35111173 Free PMC article.
-
The comparative transcriptome analysis of two green super rice genotypes with varying tolerance to salt stress.Mol Biol Rep. 2023 Dec 18;51(1):22. doi: 10.1007/s11033-023-08998-x. Mol Biol Rep. 2023. PMID: 38110786
References
-
- Baloglu M. Genome-wide in silico identification and comparison of growth regulating factor (GRF) genes in Cucurbitaceae family. Plant Omi J. 2014;7:260–270.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

