ZBED2 is an antagonist of interferon regulatory factor 1 and modifies cell identity in pancreatic cancer
- PMID: 32385160
- PMCID: PMC7261015
- DOI: 10.1073/pnas.1921484117
ZBED2 is an antagonist of interferon regulatory factor 1 and modifies cell identity in pancreatic cancer
Abstract
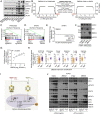
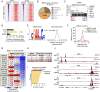
Lineage plasticity is a prominent feature of pancreatic ductal adenocarcinoma (PDA) cells, which can occur via deregulation of lineage-specifying transcription factors. Here, we show that the zinc finger protein ZBED2 is aberrantly expressed in PDA and alters tumor cell identity in this disease. Unexpectedly, our epigenomic experiments reveal that ZBED2 is a sequence-specific transcriptional repressor of IFN-stimulated genes, which occurs through antagonism of IFN regulatory factor 1 (IRF1)-mediated transcriptional activation at cooccupied promoter elements. Consequently, ZBED2 attenuates the transcriptional output and growth arrest phenotypes downstream of IFN signaling in multiple PDA cell line models. We also found that ZBED2 is preferentially expressed in the squamous molecular subtype of human PDA, in association with inferior patient survival outcomes. Consistent with this observation, we show that ZBED2 can repress the pancreatic progenitor transcriptional program, enhance motility, and promote invasion in PDA cells. Collectively, our findings suggest that high ZBED2 expression is acquired during PDA progression to suppress the IFN response pathway and to promote lineage plasticity in this disease.
Keywords: IRF1; ZBED2; interferon; lineage plasticity; pancreatic ductal adenocarcinoma.
Conflict of interest statement
Competing interest statement: C.R.V. has received funding from Boehringer-Ingelheim and is an advisor to KSQ Therapeutics.
Figures




References
-
- Grippo P. J., Nowlin P. S., Demeure M. J., Longnecker D. S., Sandgren E. P., Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 63, 2016–2019 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

