A phenome-wide association and Mendelian Randomisation study of polygenic risk for depression in UK Biobank
- PMID: 32385265
- PMCID: PMC7210889
- DOI: 10.1038/s41467-020-16022-0
A phenome-wide association and Mendelian Randomisation study of polygenic risk for depression in UK Biobank
Abstract
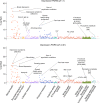
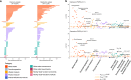
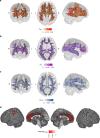
Depression is a leading cause of worldwide disability but there remains considerable uncertainty regarding its neural and behavioural associations. Here, using non-overlapping Psychiatric Genomics Consortium (PGC) datasets as a reference, we estimate polygenic risk scores for depression (depression-PRS) in a discovery (N = 10,674) and replication (N = 11,214) imaging sample from UK Biobank. We report 77 traits that are significantly associated with depression-PRS, in both discovery and replication analyses. Mendelian Randomisation analysis supports a potential causal effect of liability to depression on brain white matter microstructure (β: 0.125 to 0.868, pFDR < 0.043). Several behavioural traits are also associated with depression-PRS (β: 0.014 to 0.180, pFDR: 0.049 to 1.28 × 10-14) and we find a significant and positive interaction between depression-PRS and adverse environmental exposures on mental health outcomes. This study reveals replicable associations between depression-PRS and white matter microstructure. Our results indicate that white matter microstructure differences may be a causal consequence of liability to depression.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- World Health Organization. Depression and Other Common Mental Disorders: Global Health Estimates (World Health Organization, 2017).
Publication types
MeSH terms
Grants and funding
- 104036/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- 213674/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MR/P005748/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_17209/MRC_/Medical Research Council/United Kingdom
- G0700704/MRC_/Medical Research Council/United Kingdom
- G0200243/MRC_/Medical Research Council/United Kingdom
- U01 MH109528/MH/NIMH NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- MR/T04604X/1/MRC_/Medical Research Council/United Kingdom
- G0801418/MRC_/Medical Research Council/United Kingdom
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- MR/L010305/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

