Escherichia coli Small Proteome
- PMID: 32385980
- PMCID: PMC7212919
- DOI: 10.1128/ecosalplus.ESP-0031-2019
Escherichia coli Small Proteome
Abstract
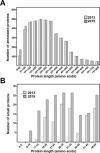
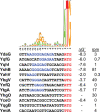
Escherichia coli was one of the first species to have its genome sequenced and remains one of the best-characterized model organisms. Thus, it is perhaps surprising that recent studies have shown that a substantial number of genes have been overlooked. Genes encoding more than 140 small proteins, defined as those containing 50 or fewer amino acids, have been identified in E. coli in the past 10 years, and there is substantial evidence indicating that many more remain to be discovered. This review covers the methods that have been successful in identifying small proteins and the short open reading frames that encode them. The small proteins that have been functionally characterized to date in this model organism are also discussed. It is hoped that the review, along with the associated databases of known as well as predicted but undetected small proteins, will aid in and provide a roadmap for the continued identification and characterization of these proteins in E. coli as well as other bacteria.
Figures

References
-
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 10.1126/science.277.5331.1453. - DOI - PubMed
-
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39(Database):D583–D590 10.1093/nar/gkq1143. [PubMed] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

