Predictive Utility of Polygenic Risk Scores for Coronary Heart Disease in Three Major Racial and Ethnic Groups
- PMID: 32386537
- PMCID: PMC7212267
- DOI: 10.1016/j.ajhg.2020.04.002
Predictive Utility of Polygenic Risk Scores for Coronary Heart Disease in Three Major Racial and Ethnic Groups
Abstract
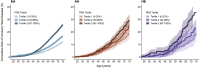
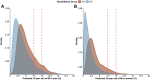
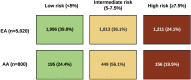
Because polygenic risk scores (PRSs) for coronary heart disease (CHD) are derived from mainly European ancestry (EA) cohorts, their validity in African ancestry (AA) and Hispanic ethnicity (HE) individuals is unclear. We investigated associations of "restricted" and genome-wide PRSs with CHD in three major racial and ethnic groups in the U.S. The eMERGE cohort (mean age 48 ± 14 years, 58% female) included 45,645 EA, 7,597 AA, and 2,493 HE individuals. We assessed two restricted PRSs (PRSTikkanen and PRSTada; 28 and 50 variants, respectively) and two genome-wide PRSs (PRSmetaGRS and PRSLDPred; 1.7 M and 6.6 M variants, respectively) derived from EA cohorts. Over a median follow-up of 11.1 years, 2,652 incident CHD events occurred. Hazard and odds ratios for the association of PRSs with CHD were similar in EA and HE cohorts but lower in AA cohorts. Genome-wide PRSs were more strongly associated with CHD than restricted PRSs were. PRSmetaGRS, the best performing PRS, was associated with CHD in all three cohorts; hazard ratios (95% CI) per 1 SD increase were 1.53 (1.46-1.60), 1.53 (1.23-1.90), and 1.27 (1.13-1.43) for incident CHD in EA, HE, and AA individuals, respectively. The hazard ratios were comparable in the EA and HE cohorts (pinteraction = 0.77) but were significantly attenuated in AA individuals (pinteraction= 2.9 × 10-3). These results highlight the potential clinical utility of PRSs for CHD as well as the need to assemble diverse cohorts to generate ancestry- and ethnicity PRSs.
Keywords: African American; coronary artery disease; coronary heart disease; genome-wide polygenic score; hispanic; ischemic heart disease; multiethnic; polygenic risk scores; risk prediction.
Copyright © 2020 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- McPherson R., Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ. Res. 2016;118:564–578. - PubMed
-
- Kullo I.J., Ding K. Mechanisms of disease: The genetic basis of coronary heart disease. Nat. Clin. Pract. Cardiovasc. Med. 2007;4:558–569. - PubMed
-
- Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HG006385/HG/NHGRI NIH HHS/United States
- U01 HG008676/HG/NHGRI NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- U01 HG006382/HG/NHGRI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- U01 HG006380/HG/NHGRI NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- UL1 TR002377/TR/NCATS NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- K24 HL137010/HL/NHLBI NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

