The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas
- PMID: 32386544
- PMCID: PMC8152789
- DOI: 10.1016/j.cell.2020.04.007
The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas
Abstract
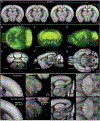
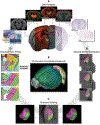
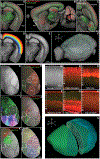
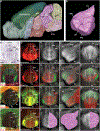
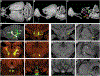
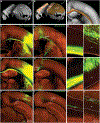
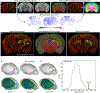
Recent large-scale collaborations are generating major surveys of cell types and connections in the mouse brain, collecting large amounts of data across modalities, spatial scales, and brain areas. Successful integration of these data requires a standard 3D reference atlas. Here, we present the Allen Mouse Brain Common Coordinate Framework (CCFv3) as such a resource. We constructed an average template brain at 10 μm voxel resolution by interpolating high resolution in-plane serial two-photon tomography images with 100 μm z-sampling from 1,675 young adult C57BL/6J mice. Then, using multimodal reference data, we parcellated the entire brain directly in 3D, labeling every voxel with a brain structure spanning 43 isocortical areas and their layers, 329 subcortical gray matter structures, 81 fiber tracts, and 8 ventricular structures. CCFv3 can be used to analyze, visualize, and integrate multimodal and multiscale datasets in 3D and is openly accessible (https://atlas.brain-map.org/).
Keywords: 3D brain atlas; CCFv3; average mouse brain; brain anatomy; brain parcellation; common coordinate framework; fiber tracts; mouse cortex; reference atlas; transgenic mice.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests D.F. is an employee Tableau Software. B.B. is an employee of 343 Industries. A.S. is an employee of Microsoft Corporation. S.W.O. is an employee of MDimune (Korea).
Figures
References
-
- Ali AA, Dale AM, Badea A, and Johnson GA (2005). Automated segmentation of neuroanatomical structures in multispectral MR microscopy of the mouse brain. Neuroimage 27, 425–435. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

