Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia
- PMID: 32386594
- PMCID: PMC7305996
- DOI: 10.1016/j.chembiol.2020.04.002
Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia
Abstract
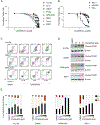
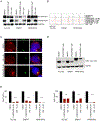
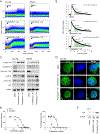
The identification of SERCA (sarco/endoplasmic reticulum calcium ATPase) as a target for modulating gain-of-function NOTCH1 mutations in Notch-dependent cancers has spurred the development of this compound class for cancer therapeutics. Despite the innate toxicity challenge associated with SERCA inhibition, we identified CAD204520, a small molecule with better drug-like properties and reduced off-target Ca2+ toxicity compared with the SERCA inhibitor thapsigargin. In this work, we describe the properties and complex structure of CAD204520 and show that CAD204520 preferentially targets mutated over wild-type NOTCH1 proteins in T cell acute lymphoblastic leukemia (T-ALL) and mantle cell lymphoma (MCL). Uniquely among SERCA inhibitors, CAD204520 suppresses NOTCH1-mutated leukemic cells in a T-ALL xenografted model without causing cardiac toxicity. This study supports the development of SERCA inhibitors for Notch-dependent cancers and extends their application to cases with isolated mutations in the PEST degradation domain of NOTCH1, such as MCL or chronic lymphocytic leukemia (CLL).
Keywords: CAD204520; NOTCH1; NOTCH1 mutation; P-type ATPases screening; PEST mutation; SERCA; T cell acute lymphoblastic leukemia (T-ALL); crystal structure; mantle cell lymphoma (MCL); thapsigargin.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Author Contributions Conceptualization and Design, M.M., A.G., W.D.-B., A.-M.L.W., M.B., and G.R.; Methodology, all authors; Resources, all authors; Investigation, all authors; Formal Analysis, all authors; Writing – Original Draft, W.D.-B., A.-M.L.W., M.M., M.B., and G.R.; Writing – Review & Editing, W.D.-B., A.-M.L.W., M.M., M.B., and G.R.; Data Curation, W.D.-B., A.-M.L.W., P.S., R.L.S., C.M., F.A., K.S., M.B., and G.R. Funding Acquisition, P.S., C.M., R.L.S., K.S., M.M., and G.R.; Project Administration, M.M. and G.R.; Supervision, G.R. Declaration of Interests W.D.-B. and A.-M.L.W. are CaDo Biotechnology IvS employees. The compounds studied are part of a patent application wholly owned by CaDo Biotechnology IvS, Denmark. K.S. has previously consulted for Novartis and Rigel Pharmaceuticals and currently receives grant funding from Novartis on topics unrelated to this manuscript.
Figures




Similar articles
-
CAD204520 Targets NOTCH1 PEST Domain Mutations in Lymphoproliferative Disorders.Int J Mol Sci. 2024 Jan 7;25(2):766. doi: 10.3390/ijms25020766. Int J Mol Sci. 2024. PMID: 38255842 Free PMC article.
-
Targeting oncogenic Notch signaling with SERCA inhibitors.J Hematol Oncol. 2021 Jan 6;14(1):8. doi: 10.1186/s13045-020-01015-9. J Hematol Oncol. 2021. PMID: 33407740 Free PMC article. Review.
-
Complementary genomic screens identify SERCA as a therapeutic target in NOTCH1 mutated cancer.Cancer Cell. 2013 Mar 18;23(3):390-405. doi: 10.1016/j.ccr.2013.01.015. Epub 2013 Feb 21. Cancer Cell. 2013. PMID: 23434461 Free PMC article.
-
Leukemia-specific delivery of mutant NOTCH1 targeted therapy.J Exp Med. 2018 Jan 2;215(1):197-216. doi: 10.1084/jem.20151778. Epub 2017 Nov 20. J Exp Med. 2018. PMID: 29158376 Free PMC article.
-
Recent advances on NOTCH signaling in T-ALL.Curr Top Microbiol Immunol. 2012;360:163-82. doi: 10.1007/82_2012_232. Curr Top Microbiol Immunol. 2012. PMID: 22673746 Review.
Cited by
-
Bcor loss promotes Richter transformation of chronic lymphocytic leukemia associated with Notch1 activation in mice.Leukemia. 2025 May;39(5):1157-1168. doi: 10.1038/s41375-025-02557-y. Epub 2025 Mar 20. Leukemia. 2025. PMID: 40113912 Free PMC article.
-
Discovery of New Anti-Cancer Agents against Patient-Derived Sorafenib-Resistant Papillary Thyroid Cancer.Int J Mol Sci. 2023 Nov 16;24(22):16413. doi: 10.3390/ijms242216413. Int J Mol Sci. 2023. PMID: 38003602 Free PMC article.
-
New Small-Molecule SERCA Inhibitors Enhance Treatment Efficacy in Lenvatinib-Resistant Papillary Thyroid Cancer.Int J Mol Sci. 2024 Oct 3;25(19):10646. doi: 10.3390/ijms251910646. Int J Mol Sci. 2024. PMID: 39408974 Free PMC article.
-
Notch signaling pathway: architecture, disease, and therapeutics.Signal Transduct Target Ther. 2022 Mar 24;7(1):95. doi: 10.1038/s41392-022-00934-y. Signal Transduct Target Ther. 2022. PMID: 35332121 Free PMC article. Review.
-
[The role and research progress of NOTCH1 in T-cell acute lymphoblastic leukemia].Zhonghua Xue Ye Xue Za Zhi. 2021 Feb 14;42(2):165-170. doi: 10.3760/cma.j.issn.0253-2727.2021.02.014. Zhonghua Xue Ye Xue Za Zhi. 2021. PMID: 33858050 Free PMC article. Chinese. No abstract available.
References
-
- Andersen JP, Lassen K, and Moller JV (1985). Changes in Ca2+ affinity related to conformational transitions in the phosphorylated state of soluble monomeric Ca2+-ATPase from sarcoplasmic reticulum. J Biol Chem 260, 371–380. - PubMed
-
- Arbabian A, Brouland JP, Gelebart P, Kovacs T, Bobe R, Enouf J, and Papp B (2011). Endoplasmic reticulum calcium pumps and cancer. Biofactors 37, 139–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

