Identification of two novel mutations in POU4F3 gene associated with autosomal dominant hearing loss in Chinese families
- PMID: 32390314
- PMCID: PMC7299729
- DOI: 10.1111/jcmm.15359
Identification of two novel mutations in POU4F3 gene associated with autosomal dominant hearing loss in Chinese families
Abstract
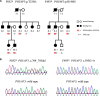
Autosomal dominant non-syndromic hearing loss is genetically heterogeneous with 47 genes identified to date, including POU4F3. In this study, by using a next-generation sequencing panel targeting 127 deafness genes, we identified a pathogenic frameshift mutation c.704_705del and a missense mutation c.593G>A in two three-generation Chinese families with late-onset progressive ADNSHL, respectively. The novel mutations of POU4F3 co-segregated with the deafness phenotype in these two families. c.704_705del caused a frameshift p.T235fs and c.593G>A caused an amino acid substitution of p.R198H. Both mutations led to an abnormal and incomplete protein structure. POU4F3 with either of the two mutations was transiently transfected into HEI-OC1 and HEK 293 cell lines and immunofluorescence assay was performed to investigate the subcellular localization of mutated protein. The results indicated that both c.704_705del (p.T235fs) and c.593G>A (p.R198H) could impair the nuclear localization function of POU4F3. The p.R198H POU4F3 protein was detected as a weak band of the correct molecular weight, indicating that the stability of p.R198H POU4F3 differed from that of the wild-type protein. While, the p.T235fs POU4F3 protein was expressed with a smaller molecular weight, implying this mutation result in a frameshift and premature termination of the POU4F3 protein. In summary, we report two novel mutations of POU4F3 associated with progressive ADNSHL and explored their effects on POU4F3 nuclear localization. These findings expanded the mutation spectrum of POU4F3 and provided new knowledge for the pathogenesis of POU4F3 in hearing loss.
Keywords: Chinese family; POU4F3; autosomal dominant; hearing loss; identification; novel mutation.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16‐31. - PubMed
-
- Morton CC, Nance WE. Current concepts: newborn hearing screening ‐ A silent revolution. N Engl J Med. 2006;354:2151‐2164. - PubMed
-
- Stelma F, Bhutta MF. Non‐syndromic hereditary sensorineural hearing loss: review of the genes involved. J Laryngol Otol. 2014;128:13‐21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

