Long-term stability of microbiome diversity and composition in fecal samples stored in eNAT medium
- PMID: 32390344
- PMCID: PMC7349174
- DOI: 10.1002/mbo3.1046
Long-term stability of microbiome diversity and composition in fecal samples stored in eNAT medium
Abstract
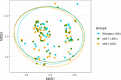
Fecal samples collected for microbiome analyses are typically frozen to avoid postcollection changes in microbial composition. eNAT is a guanidine thiocyanate-based medium that stabilizes microbial DNA and allows safe specimen handling and shipping by inactivating microorganisms. We collected fecal samples (n = 50) from children undergoing hematopoietic stem cell transplantation. We divided samples into three aliquots: (a) stored in RNAlater and immediately transferred to -80°C; (b) stored in eNAT medium and immediately transferred to -80°C; and (c) stored in eNAT medium at ambient temperature (~20°C) for 30 days prior to transfer to -80°C. Mean (standard deviation) Shannon diversity and Chao1 indices in sample aliquots were 2.05 (0.62) and 23.8 (16.6), respectively. Comparing samples frozen immediately in RNAlater to samples frozen immediately in eNAT, there were no differences in Shannon diversity (p = .51), Chao1 richness (p = .66), and overall microbiome composition (p = .99). Comparing eNAT samples frozen immediately to samples stored at ambient temperature, we identified no differences in Shannon diversity (p = .65), Chao1 richness (p = .87), and overall microbiome composition (p = .99). Storage of fecal samples in eNAT at ambient temperature for 30 days did not alter microbiome richness, diversity, or composition. eNAT may be a useful medium for fecal microbiome studies, particularly when cold chain storage is unavailable.
Keywords: 16S rRNA sequencing; bacterial inactivation; biological sample shipping; stabilization media.
© 2020 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures



References
-
- Altman, D. , & Bland, J. (1983). Measurement in medicine: The analysis of method comparison studies. The Statistician, 32(3), 307–317. 10.2307/2987937 - DOI
-
- Bentley, A. C. (2007). Shipping and handling of natural history specimens in dangerous goods. Collect Forum, 22, 66–73.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

