Biodiversity between sand grains: Meiofauna composition across southern and western Sweden assessed by metabarcoding
- PMID: 32390756
- PMCID: PMC7198628
- DOI: 10.3897/BDJ.8.e51813
Biodiversity between sand grains: Meiofauna composition across southern and western Sweden assessed by metabarcoding
Abstract
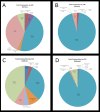
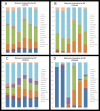
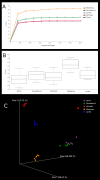
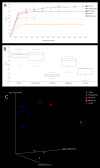
The meiofauna is an important part of the marine ecosystem, but its composition and distribution patterns are relatively unexplored. Here we assessed the biodiversity and community structure of meiofauna from five locations on the Swedish western and southern coasts using a high-throughput DNA sequencing (metabarcoding) approach. The mitochondrial cytochrome oxidase 1 (COI) mini-barcode and nuclear 18S small ribosomal subunit (18S) V1-V2 region were amplified and sequenced using Illumina MiSeq technology. Our analyses revealed a higher number of species than previously found in other areas: thirteen samples comprising 6.5 dm3 sediment revealed 708 COI and 1,639 18S metazoan OTUs. Across all sites, the majority of the metazoan biodiversity was assigned to Arthropoda, Nematoda and Platyhelminthes. Alpha and beta diversity measurements showed that community composition differed significantly amongst sites. OTUs initially assigned to Acoela, Gastrotricha and the two Platyhelminthes sub-groups Macrostomorpha and Rhabdocoela were further investigated and assigned to species using a phylogeny-based taxonomy approach. Our results demonstrate that there is great potential for discovery of new meiofauna species even in some of the most extensively studied locations.
Keywords: Acoela; Gastrotricha; Macrostomorpha; Platyhelminthes; Rhabdocoela; High Throughput Sequencing; Illumina Mi-Seq; Species Identification.
Sarah Atherton, Ulf Jondelius.
Figures

References
-
- Aller R., Aller J. Meiofauna and solute transport in marine muds. Limnology and Oceanography. 1992;37:1018–1033. doi: 10.4319/lo.1992.37.5.1018. - DOI
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Anslan Sten, Tedersoo Leho. Performance of cytochrome c oxidase subunit I (COI), ribosomal DNA Large Subunit (LSU) and Internal Transcribed Spacer 2 (ITS2) in DNA barcoding of Collembola. European Journal of Soil Biology. 2015;69:1–7. doi: 10.1016/j.ejsobi.2015.04.001. - DOI
-
- Atherton Sarah, Jondelius Ulf. Microstomum (Platyhelminthes, Macrostomorpha, Microstomidae) from the Swedish west coast: two new species and a population description. European Journal of Taxonomy. 2018;398 doi: 10.5852/ejt.2018.398. - DOI
-
- Atherton Sarah, Jondelius Ulf. Wide distributions and cryptic diversity within a Microstomum (Platyhelminthes) species complex. Zoologica Scripta. 2018;47(4):486–498. doi: 10.1111/zsc.12290. - DOI
LinkOut - more resources
Full Text Sources
