Genetic Diversity of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) From 1996 to 2017 in China
- PMID: 32390968
- PMCID: PMC7193098
- DOI: 10.3389/fmicb.2020.00618
Genetic Diversity of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) From 1996 to 2017 in China
Abstract
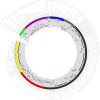
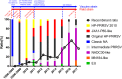
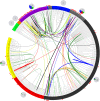
Porcine reproductive and respiratory syndrome (PRRS) is one of the most devastating diseases of the global swine industry. The causative agent porcine reproductive and respiratory syndrome virus (PRRSV) was first isolated in China in 1996 and has evolved quickly during the last two decades. To fully understand virus diversity, epidemic situation in the field, and make future predictions, a total of 365 PRRSV strains were used for evolution and genome analysis in which 353 strains were isolated from mainland China. The results showed that high diversity was found among PRRSV isolates. Total PRRSV isolates could be divided into eight subgroups. Among these subgroups strains, Original HP-PRRSV, NADC30-like, and Intermediate PRRSV were the major epidemic PRRSV strains circling in the field and would play a major role in PRRS epidemic in the future. Deletions, insertions, and recombinations have occurred frequently in the PRRSV genome. Deletions were the main driving force of viral evolution before 2006 and may also contribute further to the virus' evolution in a relatively closed or low strain diversity circumstance. The recombinant strains could be divided into three groups: the Inner group, Extensional group, and Propagating group. The evolutionary directions of the isolates in the Extensional and Propagating groups have changed, and the routes of recombination in the Propagating group were analyzed and sorted into three types. The increases in recombinant strains and high rates of recombination in recent years indicate that recombination has played a very important role in the virus' evolution. Isolates, which incorporate the advantages of their parental strains, will influence PRRSV evolution and make adverse effects on PRRS control in the future.
Keywords: evolution; genetic diversity; phylogenetic trees; porcine reproductive and respiratory syndrome virus (PRRSV); recombination.
Copyright © 2020 Jiang, Li, Yu, Li, Zhang, Zhou, Tong, Liu, Gao and Tong.
Figures


Similar articles
-
Isolation and pathogenicity comparison of two novel natural recombinant porcine reproductive and respiratory syndrome viruses with different recombination patterns in Southwest China.Microbiol Spectr. 2024 May 2;12(5):e0407123. doi: 10.1128/spectrum.04071-23. Epub 2024 Mar 21. Microbiol Spectr. 2024. PMID: 38511956 Free PMC article.
-
Molecular Characteristics and Pathogenicity of a Novel Recombinant Porcine Reproductive and Respiratory Syndrome Virus Strain from NADC30-, NADC34-, and JXA1-Like Strains That Emerged in China.Microbiol Spectr. 2022 Dec 21;10(6):e0266722. doi: 10.1128/spectrum.02667-22. Epub 2022 Nov 10. Microbiol Spectr. 2022. PMID: 36354339 Free PMC article.
-
Evolutionary analysis of six isolates of porcine reproductive and respiratory syndrome virus from a single pig farm: MLV-evolved and recombinant viruses.Infect Genet Evol. 2018 Dec;66:111-119. doi: 10.1016/j.meegid.2018.09.024. Epub 2018 Sep 24. Infect Genet Evol. 2018. PMID: 30261264
-
The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective.Virol J. 2018 Jan 4;15(1):2. doi: 10.1186/s12985-017-0910-6. Virol J. 2018. PMID: 29301547 Free PMC article. Review.
-
Porcine reproductive and respiratory syndrome virus genetic variability a management and diagnostic dilemma.Virol J. 2021 Oct 18;18(1):206. doi: 10.1186/s12985-021-01675-0. Virol J. 2021. PMID: 34663367 Free PMC article. Review.
Cited by
-
Nanopore-Based Direct RNA-Sequencing Reveals a High-Resolution Transcriptional Landscape of Porcine Reproductive and Respiratory Syndrome Virus.Viruses. 2021 Dec 16;13(12):2531. doi: 10.3390/v13122531. Viruses. 2021. PMID: 34960801 Free PMC article.
-
Advanced Research in Porcine Reproductive and Respiratory Syndrome Virus Co-infection With Other Pathogens in Swine.Front Vet Sci. 2021 Aug 26;8:699561. doi: 10.3389/fvets.2021.699561. eCollection 2021. Front Vet Sci. 2021. PMID: 34513970 Free PMC article. Review.
-
Evaluation of Four Commercial Vaccines for the Protection of Piglets against the Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus (hp-PRRSV) QH-08 Strain.Vaccines (Basel). 2021 Sep 14;9(9):1020. doi: 10.3390/vaccines9091020. Vaccines (Basel). 2021. PMID: 34579257 Free PMC article.
-
An Expeditious Neutralization Assay for Porcine Reproductive and Respiratory Syndrome Virus Based on a Recombinant Virus Expressing Green Fluorescent Protein.Curr Issues Mol Biol. 2024 Jan 23;46(2):1047-1063. doi: 10.3390/cimb46020066. Curr Issues Mol Biol. 2024. PMID: 38392184 Free PMC article.
-
Phylogenetic analysis of porcine reproductive and respiratory syndrome virus in Vietnam, 2021.Virus Genes. 2022 Aug;58(4):361-366. doi: 10.1007/s11262-022-01912-w. Epub 2022 May 19. Virus Genes. 2022. PMID: 35589912 Free PMC article.
References
-
- Brockmeier S. L., Loving C. L., Vorwald A. C., Kehrli M. E., Jr., Baker R. B., Nicholson T. L., et al. . (2012). Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 169, 212–221. 10.1016/j.virusres.2012.07.030 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

