Tree Root Zone Microbiome: Exploring the Magnitude of Environmental Conditions and Host Tree Impact
- PMID: 32390986
- PMCID: PMC7190799
- DOI: 10.3389/fmicb.2020.00749
Tree Root Zone Microbiome: Exploring the Magnitude of Environmental Conditions and Host Tree Impact
Abstract
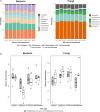
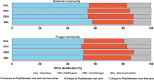
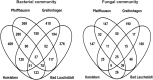
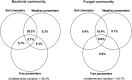
Tree roots attract their associated microbial partners from the local soil community. Accordingly, tree root-associated microbial communities are shaped by both the host tree and local environmental variables. To rationally compare the magnitude of environmental conditions and host tree impact, the "PhytOakmeter" project planted clonal oak saplings (Quercus robur L., clone DF159) as phytometers into different field sites that are within a close geographic space across the Central German lowland region. The PhytOakmeters were produced via micro-propagation to maintain their genetic identity. The current study analyzed the microbial communities in the PhytOakmeter root zone vs. the tree root-free zone of soil two years after out-planting the trees. Soil DNA was extracted, 16S and ITS2 genes were respectively amplified for bacteria and fungi, and sequenced using Illumina MiSeq technology. The obtained microbial communities were analyzed in relation to soil chemistry and weather data as environmental conditions, and the host tree growth. Although microbial diversity in soils of the tree root zone was similar among the field sites, the community structure was site-specific. Likewise, within respective sites, the microbial diversity between PhytOakmeter root and root-free zones was comparable. The number of microbial species exclusive to either zone, however, was higher in the host tree root zone than in the tree root-free zone. PhytOakmeter "core" and "site-specific" microbiomes were identified and attributed to the host tree selection effect and/or to the ambient conditions of the sites, respectively. The identified PhytOakmeter root zone-associated microbiome predominantly included ectomycorrhizal fungi, yeasts and saprotrophs. Soil pH, soil organic matter, and soil temperature were significantly correlated with the microbial diversity and/or community structure. Although the host tree contributed to shape the soil microbial communities, its effect was surpassed by the impact of environmental factors. The current study helps to understand site-specific microbe recruitment processes by young host trees.
Keywords: PhytOakmeter; core and site-specific microbiomes; environmental conditions; microbial diversity; microbial recruitment.
Copyright © 2020 Habiyaremye, Goldmann, Reitz, Herrmann and Buscot.
Figures



References
-
- Aislabie J., Deslippe J. R., Dymond J. (2013). “Soil microbes and their contribution to soil services,” in Ecosystem Services in New Zealand–Conditions and Trends, ed. Dymond J. R. (Lincoln: Manaaki Whenua Press; ), 143–161.
-
- Akinwande M. O., Dikko H. G., Samson A. (2015). Variance inflation factor: as a condition for the inclusion of suppressor variable (s) in regression analysis. Open J. Stat. 5 754–767. 10.4236/ojs.2015.57075 - DOI
-
- Anderson J. M. (1992). “Responses of soils to climate change,” in Advances in Ecological Research, eds Begon M., Fitter A. H., Macfadyen A. (Cambridge, MA: Academic Press; ), 163–210. 10.1016/S0065-2504(08)60136-1 - DOI
LinkOut - more resources
Full Text Sources

