Transient Expression of IL-17A in Foxp3 Fate-Tracked Cells in Porphyromonas gingivalis-Mediated Oral Dysbiosis
- PMID: 32391008
- PMCID: PMC7190800
- DOI: 10.3389/fimmu.2020.00677
Transient Expression of IL-17A in Foxp3 Fate-Tracked Cells in Porphyromonas gingivalis-Mediated Oral Dysbiosis
Abstract
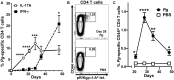
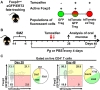
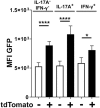
In periodontitis Porphyromonas gingivalis contributes to the development of a dysbiotic oral microbiome. This altered ecosystem elicits a diverse innate and adaptive immune response that simultaneously involves Th1, Th17, and Treg cells. It has been shown that Th17 cells can alter their gene expression to produce interferon-gamma (IFN-γ). Forkhead box P3 (Foxp3) is considered the master regulator of Treg cells that produce inhibitory cytokines like IL-10. Differentiation pathways that lead to Th17 and Treg cells from naïve progenitors are considered antagonistic. However, it has been reported that Treg cells expressing IL-17A as well as IFN-γ producing Th17 cells have been observed in several inflammatory conditions. Each scenario appears plausible with T cell transdifferentiation resulting from persistent microbial challenge and consequent inflammation. We established that oral colonization with P. gingivalis drives an initial IL-17A dominated Th17 response in the oral mucosa that is dependent on intraepithelial Langerhans cells (LCs). We hypothesized that Treg cells contribute to this initial IL-17A response through transient expression of IL-17A and that persistent mucosal colonization with P. gingivalis drives Th17 cells toward an IFN-γ phenotype at later stages of infection. We utilized fate-tracking mice where IL-17A- or Foxp3-promoter activity drives the permanent expression of red fluorescent protein tdTomato to test our hypothesis. At day 28 of infection timeline, Th17 cells dominated in the oral mucosa, outnumbering Th1 cells by 3:1. By day 48 this dominance was inverted with Th1 cells outnumbering Th17 cells by nearly 2:1. Tracking tdTomato+ Th17 cells revealed only sporadic transdifferentiation to an IFN-γ-producing phenotype by day 48; the appearance of Th1 cells at day 48 was due to a late de novo Th1 response. tdTomato+ Foxp3+ T cells were 35% of the total live CD4+T cells in the oral mucosa and 3.9% of them developed a transient IL-17A-producing phenotype by day 28. Interestingly, by day 48 these IL-17A-producing Foxp3+ T cells had disappeared. Therefore, persistent oral P. gingivalis infection stimulates an initial IL-17A-biased response led by Th17 cells and a small but significant number of IL-17A-expressing Treg cells that changes into a late de novo Th1 response with only sporadic transdifferentiation of Th17 cells.
Keywords: Foxp3; IL-17A; Porphyromonas gingivalis; Th17 cells; Treg cells; fate-tracking; periodontitis.
Copyright © 2020 Bittner-Eddy, Fischer and Costalonga.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

