The Intratumoral Heterogeneity Reflects the Intertumoral Subtypes of Glioblastoma Multiforme: A Regional Immunohistochemistry Analysis
- PMID: 32391260
- PMCID: PMC7193089
- DOI: 10.3389/fonc.2020.00494
The Intratumoral Heterogeneity Reflects the Intertumoral Subtypes of Glioblastoma Multiforme: A Regional Immunohistochemistry Analysis
Abstract
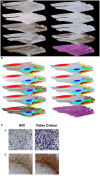
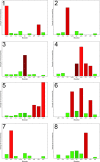
Glioblastoma multiforme (GBM) is the most frequent and aggressive primary brain tumor in adults. Despite extensive therapy the prognosis for GBM patients remains poor and the extraordinary therapy resistance has been attributed to intertumoral heterogeneity of glioblastoma. Different prognostic relevant GBM tumor subtypes have been identified based on their molecular profile. This approach, however, neglects the heterogeneity within individual tumors, that is, the intratumoral heterogeneity. Here, we detected the regional immunoreactivity by immunohistochemistry and immunofluorescence using nine different markers on resected GBM specimens (IDH wildtype, WHO grade IV). We found repetitive expression profiles, that could be classified into clusters. These clusters could then be assigned to five pathophysiologically relevant groups that reflect the previously described subclasses of GBM, including mesenchymal, classical, and proneural subtype. Our data indicate the presence of tumor differentiations and tumor subclasses that occur within individual tumors, and might therefore contribute to develop adapted, individual-based therapies.
Keywords: glioblastoma; histological architecture; histological subtypes; intertumoral heterogeneity; intratumoral heterogeneity.
Copyright © 2020 Bergmann, Delbridge, Gempt, Feuchtinger, Walch, Schirmer, Bunk, Aschenbrenner, Liesche-Starnecker and Schlegel.
Figures



References
LinkOut - more resources
Full Text Sources

