Depletion of Ric-8B leads to reduced mTORC2 activity
- PMID: 32392211
- PMCID: PMC7252638
- DOI: 10.1371/journal.pgen.1008255
Depletion of Ric-8B leads to reduced mTORC2 activity
Abstract
mTOR, a serine/threonine protein kinase that is involved in a series of critical cellular processes, can be found in two functionally distinct complexes, mTORC1 and mTORC2. In contrast to mTORC1, little is known about the mechanisms that regulate mTORC2. Here we show that mTORC2 activity is reduced in mice with a hypomorphic mutation of the Ric-8B gene. Ric-8B is a highly conserved protein that acts as a non-canonical guanine nucleotide exchange factor (GEF) for heterotrimeric Gαs/olf type subunits. We found that Ric-8B hypomorph embryos are smaller than their wild type littermates, fail to close the neural tube in the cephalic region and die during mid-embryogenesis. Comparative transcriptome analysis revealed that signaling pathways involving GPCRs and G proteins are dysregulated in the Ric-8B mutant embryos. Interestingly, this analysis also revealed an unexpected impairment of the mTOR signaling pathway. Phosphorylation of Akt at Ser473 is downregulated in the Ric-8B mutant embryos, indicating a decreased activity of mTORC2. Knockdown of the endogenous Ric-8B gene in cultured cell lines leads to reduced phosphorylation levels of Akt (Ser473), further supporting the involvement of Ric-8B in mTORC2 activity. Our results reveal a crucial role for Ric-8B in development and provide novel insights into the signals that regulate mTORC2.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
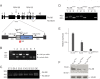

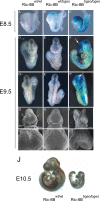
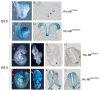
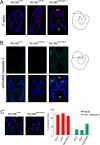
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

