Investigating gene-microRNA networks in atrial fibrillation patients with mitral valve regurgitation
- PMID: 32392228
- PMCID: PMC7213724
- DOI: 10.1371/journal.pone.0232719
Investigating gene-microRNA networks in atrial fibrillation patients with mitral valve regurgitation
Abstract
Background: Atrial fibrillation (AF) is predicted to affect around 17.9 million individuals in Europe by 2060. The disease is associated with severe electrical and structural remodelling of the heart, and increased the risk of stroke and heart failure. In order to improve treatment and find new drug targets, the field needs to better comprehend the exact molecular mechanisms in these remodelling processes.
Objectives: This study aims to identify gene and miRNA networks involved in the remodelling of AF hearts in AF patients with mitral valve regurgitation (MVR).
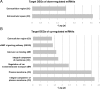
Methods: Total RNA was extracted from right atrial biopsies from patients undergoing surgery for mitral valve replacement or repair with AF and without history of AF to test for differentially expressed genes and miRNAs using RNA-sequencing and miRNA microarray. In silico predictions were used to construct a mRNA-miRNA network including differentially expressed mRNAs and miRNAs. Gene and chromosome enrichment analysis were used to identify molecular pathways and high-density AF loci.
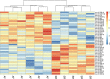
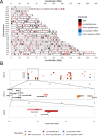
Results: We found 644 genes and 43 miRNAs differentially expressed in AF patients compared to controls. From these lists, we identified 905 pairs of putative miRNA-mRNA interactions, including 37 miRNAs and 295 genes. Of particular note, AF-associated miR-130b-3p, miR-338-5p and miR-208a-3p were differentially expressed in our AF tissue samples. These miRNAs are predicted regulators of several differentially expressed genes associated with cardiac conduction and fibrosis. We identified two high-density AF loci in chromosomes 14q11.2 and 6p21.3.
Conclusions: AF in MVR patients is associated with down-regulation of ion channel genes and up-regulation of extracellular matrix genes. Other AF related genes are dysregulated and several are predicted to be targeted by miRNAs. Our novel miRNA-mRNA regulatory network provides new insights into the mechanisms of AF.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, et al. Genome-wide association study of 1 million people identifies 111 loci for atrial fibrillation. bioRxiv. 2018. January 4;242149.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

