Ecology, Structure, and Evolution of Shigella Phages
- PMID: 32392456
- PMCID: PMC7670969
- DOI: 10.1146/annurev-virology-010320-052547
Ecology, Structure, and Evolution of Shigella Phages
Abstract
Numerous bacteriophages-viruses of bacteria, also known as phages-have been described for hundreds of bacterial species. The Gram-negative Shigella species are close relatives of Escherichia coli, yet relatively few previously described phages appear to exclusively infect this genus. Recent efforts to isolate Shigella phages have indicated these viruses are surprisingly abundant in the environment and have distinct genomic and structural properties. In addition, at least one model system used for experimental evolution studies has revealed a unique mechanism for developing faster infection cycles. Differences between these bacteriophages and other well-described model systems may mirror differences between their hosts' ecology and defense mechanisms. In this review, we discuss the history of Shigella phages and recent developments in their isolation and characterization and the structural information available for three model systems, Sf6, Sf14, and HRP29; we also provide an overview of potential selective pressures guiding both Shigella phage and host evolution.
Keywords: Omps; Shigella; lipopolysaccharide; myoviruses; phage biology; podoviruses.
Figures
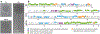
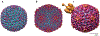
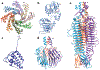
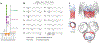
References
-
- Rautureau GJP, Palama TL, Canard I, Mirande C, Chatellier S,et al. 2019. Discrimination of Escherichia coli and Shigella spp. by nuclear magnetic resonance based metabolomic characterization of culture media. ACS Infect. Dis. 5:1879–86 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

