Targeting RGS4 Ablates Glioblastoma Proliferation
- PMID: 32392739
- PMCID: PMC7247588
- DOI: 10.3390/ijms21093300
Targeting RGS4 Ablates Glioblastoma Proliferation
Abstract
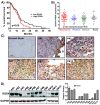
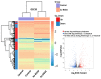
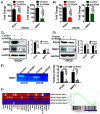
Glioblastoma (GBM) is the most common type of adult primary brain tumor with a median survival rate of less than 15 months, regardless of the current standard of care. Cellular heterogeneity, self-renewal ability and tumorigenic glioma cancer stem cell (GSC) populations contribute to the difficulty in treating GBM. G-protein-coupled receptors (GPCRs) are the largest group of membrane proteins and mediate many cellular responses. Regulators of G-protein signaling 4 (RGS4) are negative regulators of G-protein signaling, and elevated levels of RGS4 are reportedly linked with several human diseases, including cancer. This study investigates the effect of silencing RGS4, resulting in inhibition of GSC growth, invasion and migration. Data obtained from The Cancer Genome Atlas (TCGA) demonstrated poor patient survival with high expression of RGS4. Immunohistochemistry and immunoblot analysis conducted on GBM patient biopsy specimens demonstrated increased RGS4 expression correlative with the TCGA data. RNA sequencing confirmed a significant decrease in the expression of markers involved in GSC invasion and migration, particularly matrix metalloproteinase-2 (MMP2) in knockout of RGS4 using CRISPR plasmid (ko-RGS4)-treated samples compared to parental controls. Gelatin zymography confirmed the reduced activity of MMP2 in ko-RGS4-treated samples. Silencing RGS4 further reduced the invasive and migratory abilities and induction of apoptosis of GSCs as evidenced by Matrigel plug assay, wound healing assay and human apoptosis array. Collectively, our results showed that the silencing of RGS4 plays an important role in regulating multiple cellular functions, and is an important therapeutic target in GBM.
Keywords: MMP2; RGS4; apoptosis; glioma stem cells.
Conflict of interest statement
The authors declare no conflict of interest that exists with this manuscript.
Figures
References
-
- Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of Radiotherapy with Concomitant and Adjuvant Temozolomide Versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase III Study: 5-Year Analysis of the EORTC-NCIC Trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
-
- Gimple R.C., Kidwell R.L., Kim L.J.Y., Sun T., Gromovsky A.D., Wu Q., Wolf M., Lv D., Bhargava S., Jiang L., et al. Glioma Stem Cell Specific Super Enhancer Promotes Polyunsaturated Fatty Acid Synthesis to Support EGFR Signaling. Cancer Discov. 2019;9:1248–12678. doi: 10.1158/2159-8290.CD-19-0061. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

