Transgelin is a poor prognostic factor associated with advanced colorectal cancer (CRC) stage promoting tumor growth and migration in a TGFβ-dependent manner
- PMID: 32393769
- PMCID: PMC7214449
- DOI: 10.1038/s41419-020-2529-6
Transgelin is a poor prognostic factor associated with advanced colorectal cancer (CRC) stage promoting tumor growth and migration in a TGFβ-dependent manner
Abstract
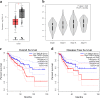
Colorectal cancer (CRC) is the fourth most common cancer type globally. Investigating the signaling pathways that maintain cancer cell phenotype can identify new biomarkers for targeted therapy. Aberrant transforming growth factor-β (TGFβ) signaling has been implicated in CRC progression, however, the exact mechanism by which TGFβ exerts its function is still being unraveled. Herein, we investigated TAGLN expression, prognostic value, and its regulation by TGFβ in CRC. While TAGLN was generally found to be downregulated in CRC, elevated expression of TAGLN was associated with advanced CRC stage and predicted poor overall survival (hazard ratio (HR) = 1.8, log-rank test P-value = 0.014) and disease-free survival (HR = 1.6, log-rank test P-value = 0.046), hence implicating TAGLN as poor prognostic factor in CRC. Forced expression of TAGLN was associated with enhanced CRC cell proliferation, clonogenic growth, cell migration and in vivo tumor formation in immunocompromised mice, while targeted depletion of TAGLN exhibited opposing biological effects. Global gene expression profiling of TAGLN-overexpressing or TAGLN-deficient CRC cell lines revealed deregulation of multiple cancer-related genes and signaling pathways. Transmission electron microscopy (TEM) revealed ultrastructural changes due to loss of TAGLN, including disruption of actin cytoskeleton organization and aberrant actin filament distribution. Hierarchical clustering, principle component, and ingenuity pathway analyses revealed distinct molecular profile associated with TAGLNhigh CRC patients with remarkable activation of a number of mechanistic networks, including SMARCA4, TGFβ1, and P38 MAPK. The P38 MAPK was the top predicted upstream regulator network promoting cell movement through regulation of several intermediate molecules, including TGFβ1. Concordantly, functional categories associated with cellular movement and angiogenesis were also enriched in TAGLNhigh CRC, supporting a model for the molecular mechanisms linking TGFβ-induced upregulation of TAGLN and CRC tumor progression and suggesting TAGLN as potential prognostic marker associated with advanced CRC pathological stage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

