Protein-altering germline mutations implicate novel genes related to lung cancer development
- PMID: 32393777
- PMCID: PMC7214407
- DOI: 10.1038/s41467-020-15905-6
Protein-altering germline mutations implicate novel genes related to lung cancer development
Abstract
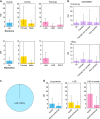
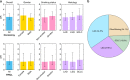
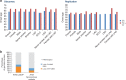
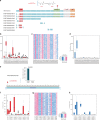
Few germline mutations are known to affect lung cancer risk. We performed analyses of rare variants from 39,146 individuals of European ancestry and investigated gene expression levels in 7,773 samples. We find a large-effect association with an ATM L2307F (rs56009889) mutation in adenocarcinoma for discovery (adjusted Odds Ratio = 8.82, P = 1.18 × 10-15) and replication (adjusted OR = 2.93, P = 2.22 × 10-3) that is more pronounced in females (adjusted OR = 6.81 and 3.19 and for discovery and replication). We observe an excess loss of heterozygosity in lung tumors among ATM L2307F allele carriers. L2307F is more frequent (4%) among Ashkenazi Jewish populations. We also observe an association in discovery (adjusted OR = 2.61, P = 7.98 × 10-22) and replication datasets (adjusted OR = 1.55, P = 0.06) with a loss-of-function mutation, Q4X (rs150665432) of an uncharacterized gene, KIAA0930. Our findings implicate germline genetic variants in ATM with lung cancer susceptibility and suggest KIAA0930 as a novel candidate gene for lung cancer risk.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA119997/CA/NCI NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- U01 CA209414/CA/NCI NIH HHS/United States
- 202849/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA111703/CA/NCI NIH HHS/United States
- UL1 TR000117/TR/NCATS NIH HHS/United States
- WT202849/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- P20 CA090578/CA/NCI NIH HHS/United States
- U19 CA148127/CA/NCI NIH HHS/United States
- P20 GM103534/GM/NIGMS NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- R01 CA092824/CA/NCI NIH HHS/United States
- R35 CA197449/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- UM1 CA164973/CA/NCI NIH HHS/United States
- U01 CA167462/CA/NCI NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
- P20 RR018787/RR/NCRR NIH HHS/United States
- DH_/Department of Health/United Kingdom
- S10 RR025141/RR/NCRR NIH HHS/United States
- R01 CA074386/CA/NCI NIH HHS/United States
- R01 CA176568/CA/NCI NIH HHS/United States
- K07 CA172294/CA/NCI NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R56 LM012371/LM/NLM NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- P01 CA033619/CA/NCI NIH HHS/United States
- P30 CA177558/CA/NCI NIH HHS/United States
- P50 CA090578/CA/NCI NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- G0902313/MRC_/Medical Research Council/United Kingdom
- 24390/CRUK_/Cancer Research UK/United Kingdom
- R01 CA151989/CA/NCI NIH HHS/United States
- UM1 CA167462/CA/NCI NIH HHS/United States
- P30 CA071789/CA/NCI NIH HHS/United States
- 001/WHO_/World Health Organization/International
- U01 CA164973/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

