Fecal microbiota transplant rescues mice from human pathogen mediated sepsis by restoring systemic immunity
- PMID: 32393794
- PMCID: PMC7214422
- DOI: 10.1038/s41467-020-15545-w
Fecal microbiota transplant rescues mice from human pathogen mediated sepsis by restoring systemic immunity
Abstract
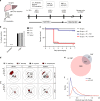
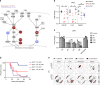
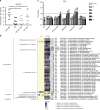
Death due to sepsis remains a persistent threat to critically ill patients confined to the intensive care unit and is characterized by colonization with multi-drug-resistant healthcare-associated pathogens. Here we report that sepsis in mice caused by a defined four-member pathogen community isolated from a patient with lethal sepsis is associated with the systemic suppression of key elements of the host transcriptome required for pathogen clearance and decreased butyrate expression. More specifically, these pathogens directly suppress interferon regulatory factor 3. Fecal microbiota transplant (FMT) reverses the course of otherwise lethal sepsis by enhancing pathogen clearance via the restoration of host immunity in an interferon regulatory factor 3-dependent manner. This protective effect is linked to the expansion of butyrate-producing Bacteroidetes. Taken together these results suggest that fecal microbiota transplantation may be a treatment option in sepsis associated with immunosuppression.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- WHO. Antimicrobial resistance: global report on surveillance 2014. WHO. http://www.who.int/drugresistance/documents/surveillancereport/en/. (Accessed: 28th June 2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

