T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers
- PMID: 32393800
- PMCID: PMC7303014
- DOI: 10.1038/s41591-020-0858-8
T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers
Abstract
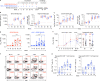
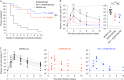
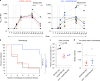
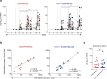
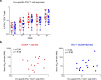
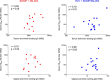
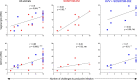
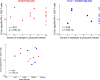
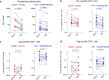
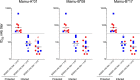
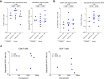
Recent efforts toward an HIV vaccine focus on inducing broadly neutralizing antibodies, but eliciting both neutralizing antibodies (nAbs) and cellular responses may be superior. Here, we immunized macaques with an HIV envelope trimer, either alone to induce nAbs, or together with a heterologous viral vector regimen to elicit nAbs and cellular immunity, including CD8+ tissue-resident memory T cells. After ten vaginal challenges with autologous virus, protection was observed in both vaccine groups at 53.3% and 66.7%, respectively. A nAb titer >300 was generally associated with protection but in the heterologous viral vector + nAb group, titers <300 were sufficient. In this group, protection was durable as the animals resisted six more challenges 5 months later. Antigen stimulation of T cells in ex vivo vaginal tissue cultures triggered antiviral responses in myeloid and CD4+ T cells. We propose that cellular immune responses reduce the threshold of nAbs required to confer superior and durable protection.
Conflict of interest statement
M.T. and J.V. are employees of 3M, the manufacturer of 3M-052 used as an adjuvant in this study.
Figures




References
-
- Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

