NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices
- PMID: 32393888
- PMCID: PMC7367149
- DOI: 10.1038/s41556-020-0513-0
NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices
Abstract
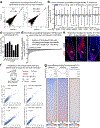
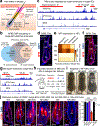
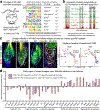
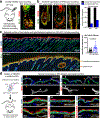
Tissue homeostasis and regeneration rely on resident stem cells (SCs), whose behaviour is regulated through niche-dependent crosstalk. The mechanisms underlying SC identity are still unfolding. Here, using spatiotemporal gene ablation in murine hair follicles, we uncover a critical role for the transcription factors (TFs) nuclear factor IB (NFIB) and IX (NFIX) in maintaining SC identity. Without NFI TFs, SCs lose their hair-regenerating capability, and produce skin bearing striking resemblance to irreversible human alopecia, which also displays reduced NFIs. Through single-cell transcriptomics, ATAC-Seq and ChIP-Seq profiling, we expose a key role for NFIB and NFIX in governing super-enhancer maintenance of the key hair follicle SC-specific TF genes. When NFIB and NFIX are genetically removed, the stemness epigenetic landscape is lost. Super-enhancers driving SC identity are decommissioned, while unwanted lineages are de-repressed ectopically. Together, our findings expose NFIB and NFIX as crucial rheostats of tissue homeostasis, functioning to safeguard the SC epigenome from a breach in lineage confinement that otherwise triggers irreversible tissue degeneration.
Conflict of interest statement
COMPETING INTERESTS
The authors declare the following competing interests: R.C.A. is currently employed at Regeneron Pharmaceuticals. E.F. is on the Scientific Advisory Board of L’Oreal and Arsenal Biosciences.
Figures
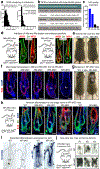
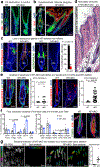


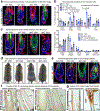
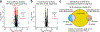





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

