Hepatic Molecular Signatures Highlight the Sexual Dimorphism of Nonalcoholic Steatohepatitis (NASH)
- PMID: 32394476
- PMCID: PMC8048532
- DOI: 10.1002/hep.31312
Hepatic Molecular Signatures Highlight the Sexual Dimorphism of Nonalcoholic Steatohepatitis (NASH)
Abstract
Background and aims: Nonalcoholic steatohepatitis (NASH) is considered as a pivotal stage in nonalcoholic fatty liver disease (NAFLD) progression, given that it paves the way for severe liver injuries such as fibrosis and cirrhosis. The etiology of human NASH is multifactorial, and identifying reliable molecular players and/or biomarkers has proven difficult. Together with the inappropriate consideration of risk factors revealed by epidemiological studies (altered glucose homeostasis, obesity, ethnicity, sex, etc.), the limited availability of representative NASH cohorts with associated liver biopsies, the gold standard for NASH diagnosis, probably explains the poor overlap between published "omics"-defined NASH signatures.
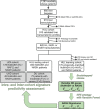
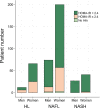
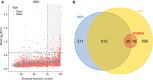
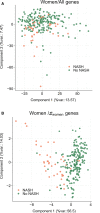
Approach and results: Here, we have explored transcriptomic profiles of livers starting from a 910-obese-patient cohort, which was further stratified based on stringent histological characterization, to define "NoNASH" and "NASH" patients. Sex was identified as the main factor for data heterogeneity in this cohort. Using powerful bootstrapping and random forest (RF) approaches, we identified reliably differentially expressed genes participating in distinct biological processes in NASH as a function of sex. RF-calculated gene signatures identified NASH patients in independent cohorts with high accuracy.
Conclusions: This large-scale analysis of transcriptomic profiles from human livers emphasized the sexually dimorphic nature of NASH and its link with fibrosis, calling for the integration of sex as a major determinant of liver responses to NASH progression and responses to drugs.
© 2020 The Authors. Hepatology published by Wiley Periodicals LLC on behalf of American Association for the Study of Liver Diseases.
Figures




References
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11‐20. - PubMed
-
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73‐84. - PubMed
-
- Argo CK, Northup PG, Al‐Osaimi AM, Caldwell SH. Systematic review of risk factors for fibrosis progression in non‐alcoholic steatohepatitis. J Hepatol 2009;51:371‐379. - PubMed

