Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance
- PMID: 32394893
- PMCID: PMC7217694
- DOI: 10.7554/eLife.51015
Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance
Abstract
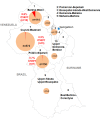
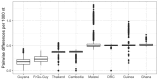
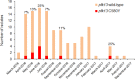
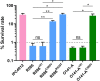
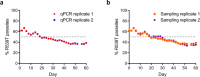
Antimalarial drug resistance has historically arisen through convergent de novo mutations in Plasmodium falciparum parasite populations in Southeast Asia and South America. For the past decade in Southeast Asia, artemisinins, the core component of first-line antimalarial therapies, have experienced delayed parasite clearance associated with several pfk13 mutations, primarily C580Y. We report that mutant pfk13 has emerged independently in Guyana, with genome analysis indicating an evolutionary origin distinct from Southeast Asia. Pfk13 C580Y parasites were observed in 1.6% (14/854) of samples collected in Guyana in 2016-2017. Introducing pfk13 C580Y or R539T mutations by gene editing into local parasites conferred high levels of in vitro artemisinin resistance. In vitro growth competition assays revealed a fitness cost associated with these pfk13 variants, potentially explaining why these resistance alleles have not increased in frequency more quickly in South America. These data place local malaria control efforts at risk in the Guiana Shield.
Keywords: Guyana; P. falciparum; South America; artemisinin resistance; epidemiology; evolution; global health; infectious disease; kelch 13; malaria; microbiology.
Plain language summary
All recommended treatments against malaria include a drug called artemisinin or some of its derivatives. However, there are concerns that Plasmodium falciparum, the parasite that causes most cases of malaria, will eventually develop widespread resistance to the drug. A strain of P. falciparum partially resistant to artemisinin was seen in Cambodia in 2008, and it has since spread across Southeast Asia. The resistance appears to be frequently linked to a mutation known as pfk13 C580Y. Southeast Asia and Amazonia are considered to be hotspots for antimalarial drug resistance, and the pfk13 C580Y mutation was detected in the South American country of Guyana in 2010. To examine whether the mutation was still circulating in this part of the world, Mathieu et al. collected and analyzed 854 samples across Guyana between 2016 and 2017. Overall, 1.6% of the samples had the pfk13 C580Y mutation, but this number was as high as 8.8% in one region. Further analyses revealed that the mutation in Guyana had not spread from Southeast Asia, but that it had occurred in Amazonia independently. To better understand the impact of the pfk13 C580Y mutation, Mathieu et al. introduced this genetic change into non-resistant parasites from a country neighbouring Guyana. As expected, the mutation made P. falciparum highly resistant to artemisinin, but it also slowed the growth rate of the parasite. This disadvantage may explain why the mutation has not spread more rapidly through Guyana in recent years. Artemisinin and its derivatives are always associated with other antimalarial drugs to slow the development of resistance; there are concerns that reduced susceptibility to artemisinin leads to the parasites becoming resistant to the partner drugs. Further research is needed to evaluate how the pfk13 C580Y mutation affects the parasite’s response to the typical combination of drugs that are given to patients.
© 2020, Mathieu et al.
Conflict of interest statement
LM, HC, AE, SM, YL, JP, NL, QG, VU, MD, DN, DF, LM No competing interests declared, MA, JA, PR MPA, JSFA, and PR are staff members of the World Health Organization. The authors alone are responsible for the views expressed in this publication and they do not necessarily represent the decisions, policy or views of the World Health Organization.
Figures



References
-
- Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, Fairhurst RM, Kwiatkowski DP. Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: a retrospective genetic study. The Lancet Infectious Diseases. 2018;18:337–345. doi: 10.1016/S1473-3099(18)30068-9. - DOI - PMC - PubMed
-
- Anderson TJ, Nair S, McDew-White M, Cheeseman IH, Nkhoma S, Bilgic F, McGready R, Ashley E, Pyae Phyo A, White NJ, Nosten F. Population parameters underlying an ongoing soft sweep in southeast asian malaria parasites. Molecular Biology and Evolution. 2017;34:131–144. doi: 10.1093/molbev/msw228. - DOI - PMC - PubMed
-
- Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. - DOI - PMC - PubMed
-
- Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ, Tracking Resistance to Artemisinin Collaboration (TRAC) Spread of artemisinin resistance in Plasmodium falciparum malaria. The New England Journal of Medicine. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- W81XWH1910086/U.S. Department of Defense/International
- U19AI110818/National Institute of Allergy and Infectious Diseases/International
- R01 AI109023/NH/NIH HHS/United States
- OPP1201387/Bill and Melinda Gates Foundation/International
- R37 AI050234/AI/NIAID NIH HHS/United States
- R01 124678/NH/NIH HHS/United States
- U19 AI110818/AI/NIAID NIH HHS/United States
- 001/WHO_/World Health Organization/International
- GY0012082/European Commission/International
- ANR-10-LABX-25-01/Agence Nationale de la Recherche/International
- R37 AI50234/NH/NIH HHS/United States
- R01 AI124678/AI/NIAID NIH HHS/United States
- R01 AI109023/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

