Mitochondrial DNA Stress Signalling Protects the Nuclear Genome
- PMID: 32395698
- PMCID: PMC7213273
- DOI: 10.1038/s42255-019-0150-8
Mitochondrial DNA Stress Signalling Protects the Nuclear Genome
Abstract
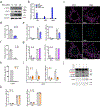
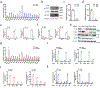
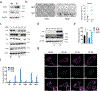
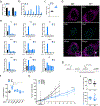
The mammalian genome comprises nuclear DNA (nDNA) derived from both parents and mitochondrial DNA (mtDNA) that is maternally inherited and encodes essential proteins required for oxidative phosphorylation. Thousands of copies of the circular mtDNA are present in most cell types that are packaged by TFAM into higher-order structures called nucleoids1. Mitochondria are also platforms for antiviral signalling2 and, due to their bacterial origin, mtDNA and other mitochondrial components trigger innate immune responses and inflammatory pathology2,3. We showed previously that instability and cytoplasmic release of mtDNA activates the cGAS-STING-TBK1 pathway resulting in interferon stimulated gene (ISG) expression that promotes antiviral immunity4. Here, we find that persistent mtDNA stress is not associated with basally activated NF-κB signalling or interferon gene expression typical of an acute antiviral response. Instead, a specific subset of ISGs, that includes Parp9, remains activated by the unphosphorylated form of ISGF3 (U-ISGF3) that enhances nDNA damage and repair responses. In cultured primary fibroblasts and cancer cells, the chemotherapeutic drug doxorubicin causes mtDNA damage and release, which leads to cGAS-STING-dependent ISG activation. In addition, mtDNA stress in TFAM-deficient mouse melanoma cells produces tumours that are more resistant to doxorubicin in vivo. Finally, Tfam +/- mice exposed to ionizing radiation exhibit enhanced nDNA repair responses in spleen. Therefore, we propose that damage to and subsequent release of mtDNA elicits a protective signalling response that enhances nDNA repair in cells and tissues, suggesting mtDNA is a genotoxic stress sentinel.
Conflict of interest statement
Competing Interests Statement. The authors have no competing interests to declare with regard to the data presented in the manuscript.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

