A de novo chromosome-level genome assembly of Coregonus sp. "Balchen": One representative of the Swiss Alpine whitefish radiation
- PMID: 32395896
- PMCID: PMC7497118
- DOI: 10.1111/1755-0998.13187
A de novo chromosome-level genome assembly of Coregonus sp. "Balchen": One representative of the Swiss Alpine whitefish radiation
Abstract
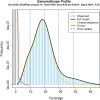
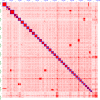
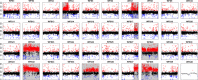
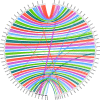
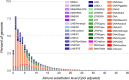
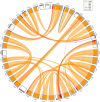
Salmonids are of particular interest to evolutionary biologists due to their incredible diversity of life-history strategies and the speed at which many salmonid species have diversified. In Switzerland alone, over 30 species of Alpine whitefish from the subfamily Coregoninae have evolved since the last glacial maximum, with species exhibiting a diverse range of morphological and behavioural phenotypes. This, combined with the whole genome duplication which occurred in the ancestor of all salmonids, makes the Alpine whitefish radiation a particularly interesting system in which to study the genetic basis of adaptation and speciation and the impacts of ploidy changes and subsequent rediploidization on genome evolution. Although well-curated genome assemblies exist for many species within Salmonidae, genomic resources for the subfamily Coregoninae are lacking. To assemble a whitefish reference genome, we carried out PacBio sequencing from one wild-caught Coregonus sp. "Balchen" from Lake Thun to ~90× coverage. PacBio reads were assembled independently using three different assemblers, falcon, canu and wtdbg2 and subsequently scaffolded with additional Hi-C data. All three assemblies were highly contiguous, had strong synteny to a previously published Coregonus linkage map, and when mapping additional short-read data to each of the assemblies, coverage was fairly even across most chromosome-scale scaffolds. Here, we present the first de novo genome assembly for the Salmonid subfamily Coregoninae. The final 2.2-Gb wtdbg2 assembly included 40 scaffolds, an N50 of 51.9 Mb and was 93.3% complete for BUSCOs. The assembly consisted of ~52% transposable elements and contained 44,525 genes.
Keywords: Alpine whitefish; Coregonus; Salmonidae; genome assembly; whitefish.
© 2020 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Figures
References
-
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Bernatchez, L. , & Dodson, J. J. (1990). Allopatric origin of sympatric populatinos of lake whitefish (Coregonus clupeaformis) as revealed by mitochondrial‐DNA restriction analysis. Evolution; International Journal of Organic Evolution, 44, 1263–1271. - PubMed
-
- Bickhart, D. M. , Rosen, B. D. , Koren, S. , Sayre, B. L. , Hastie, A. R. , Chan, S. , … Smith, T. P. L. (2017). Single‐molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nature Genetics, 49, 643–650. 10.1038/ng.3802 - DOI - PMC - PubMed
-
- Blumstein, D. M. , Campbell, M. A. , Hale, M. C. , Sutherland, B. J. G. , McKinney, G. J. , Stott, W. , & Larson, W. A. (2020). Comparative genomic analyses and a novel linkage map for cisco (Coregonus artedi) provides insight into chromosomal evolution and rediploidization across salmonids. bioRxiv, 834937, 1–35. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

