Kinetics and Optimization of the Lysine-Isopeptide Bond Forming Sortase Enzyme from Corynebacterium diphtheriae
- PMID: 32396336
- PMCID: PMC8153732
- DOI: 10.1021/acs.bioconjchem.0c00163
Kinetics and Optimization of the Lysine-Isopeptide Bond Forming Sortase Enzyme from Corynebacterium diphtheriae
Abstract
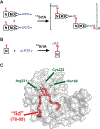
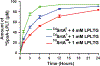
Site-specifically modified protein bioconjugates have important applications in biology, chemistry, and medicine. Functionalizing specific protein side chains with enzymes using mild reaction conditions is of significant interest, but remains challenging. Recently, the lysine-isopeptide bond forming activity of the sortase enzyme that builds surface pili in Corynebacterium diphtheriae (CdSrtA) has been reconstituted in vitro. A mutationally activated form of CdSrtA was shown to be a promising bioconjugating enzyme that can attach Leu-Pro-Leu-Thr-Gly peptide fluorophores to a specific lysine residue within the N-terminal domain of the SpaA protein (NSpaA), enabling the labeling of target proteins that are fused to NSpaA. Here we present a detailed analysis of the CdSrtA catalyzed protein labeling reaction. We show that the first step in catalysis is rate limiting, which is the formation of the CdSrtA-peptide thioacyl intermediate that subsequently reacts with a lysine ε-amine in NSpaA. This intermediate is surprisingly stable, limiting spurious proteolysis of the peptide substrate. We report the discovery of a new enzyme variant (CdSrtAΔ) that has significantly improved transpeptidation activity, because it completely lacks an inhibitory polypeptide appendage ("lid") that normally masks the active site. We show that the presence of the lid primarily impairs formation of the thioacyl intermediate and not the recognition of the NSpaA substrate. Quantitative measurements reveal that CdSrtAΔ generates its cross-linked product with a catalytic turnover number of 1.4 ± 0.004 h-1 and that it has apparent KM values of 0.16 ± 0.04 and 1.6 ± 0.3 mM for its NSpaA and peptide substrates, respectively. CdSrtAΔ is 7-fold more active than previously studied variants, labeling >90% of NSpaA with peptide within 6 h. The results of this study further improve the utility of CdSrtA as a protein labeling tool and provide insight into the enzyme catalyzed reaction that underpins protein labeling and pilus biogenesis.
Figures



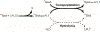
References
-
- Chudasama V; Maruani A; Caddick S, Recent advances in the construction of antibody drug conjugates. Nature Chemistry 2015, 8, (2), 114–119. - PubMed
-
- Hoyt EA; Cal PMSD; Oliveira BL; Bernardes G. a. J. L., Contemporary approaches to site-selective protein modification. Nature Reviews Chemistry 2019, 3, (3), 147–171.
-
- Elizabeth AS; Esther B; Amy EP, A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annual Review of Physiology 2016, 79, (1), 93–117. - PubMed

