A novel CpG-methylation-based nomogram predicts survival in colorectal cancer
- PMID: 32396412
- PMCID: PMC7595584
- DOI: 10.1080/15592294.2020.1762368
A novel CpG-methylation-based nomogram predicts survival in colorectal cancer
Abstract
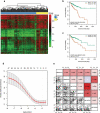
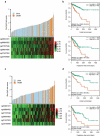
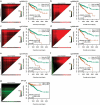
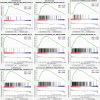
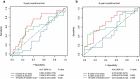
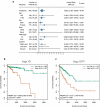
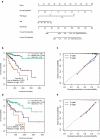
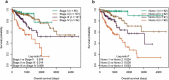
Aberrant DNA methylation is significantly associated with the prognosis of patients with colorectal cancer (CRC). Therefore, the aim of this study was to develop a CpG-methylation-based nomogram for prognostic prediction in CRC. First, 378 CRC patients with methylation data from The Cancer Genome Atlas were randomly divided into training cohort (n = 249) and test cohort (n = 129). A multistep screening strategy was performed to identify six CpG sites that were significantly associated with overall survival in the training cohort. Then, Cox regression modelling was performed to construct a prognostic signature based on the candidate CpG sites. The six-CpG signature successfully separated patients into high-risk and low-risk groups in both training and test cohorts, and its performance was superior to that of previously published methylation markers (P < 0.05). Furthermore, we established a prognostic nomogram incorporating this signature, TNM stage, and age. The nomogram exhibited better prediction for overall survival in comparison with the three independent prognostic factors in the training cohort (C-index: 0.798 vs 0.620 to 0.737; P < 0.001). In the test cohort, the performance of nomogram was also superior to that of the three independent prognostic factors (C-index: 0.715 vs 0.590 to 0.665; P < 0.05). Meanwhile, the calibration curves for survival probability showed good agreement between prediction by nomogram and actual observation in both training and test cohorts. Together, the present study provides a novel CpG-methylation-based nomogram as a promising predictor for overall survival of CRC patients, which may help improve decision-making regarding the personalized treatments of patients with CRC.
Keywords: DNA methylation; colorectal cancer; nomogram; prognosis; survival analysis.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. November;68(6):394–424. . - PubMed
-
- Guraya SY, Pattern S.. Time of recurrent colorectal cancer after curative surgery. Clin Colorectal Cancer. 2019. June;18(2):e223–e228. - PubMed
-
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006. July;38(7):787–793. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
