STAT1-induced upregulation of lncRNA KTN1-AS1 predicts poor prognosis and facilitates non-small cell lung cancer progression via miR-23b/DEPDC1 axis
- PMID: 32396871
- PMCID: PMC7244022
- DOI: 10.18632/aging.103191
STAT1-induced upregulation of lncRNA KTN1-AS1 predicts poor prognosis and facilitates non-small cell lung cancer progression via miR-23b/DEPDC1 axis
Abstract
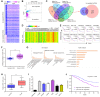
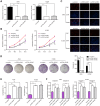
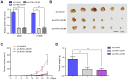
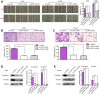
Several of the thousands of long noncoding RNAs (lncRNAs) have been functionally characterized in various tumors. In this study, we aimed to explore the function and possible molecular mechanism of lncRNA KTN1 antisense RNA 1 (KTN1-AS1) involved in non-small cell lung cancer (NSCLC). We identified a novel NSCLC-related lncRNA, KTN1 antisense RNA 1 (KTN1-AS1) which was demonstrated to be distinctly highly expressed in NSCLC. KTN1-AS1 upregulation was induced by STAT1. Clinical study also suggested that higher levels of KTN1-AS1 were associated with advanced clinical progression and a shorter five-year overall survival. Functionally, loss-of-function assays with in vitro and in vivo experiments revealed that KTN1-AS1 promoted the proliferation, migration, invasion and EMT progress of NSCLC cells, and suppressed apoptosis. Mechanistic studies indicated that miR-23b was a direct target of KTN1-AS1, which functioned as a ceRNA to subsequently facilitate miR-23b's target gene DEPDC1 expression in NSCLC cells. Rescue experiments confirmed that KTN1-AS1 overexpression could increase the colony formation and migration ability suppressed by miR-23b upregulation in NSCLC cells. Overall, our findings imply that STAT1-induced upregulation of KTN1-AS1 display tumor-promotive roles in NSCLC progression via regulating miR-23b/DEPDC1 axis, suggesting that KTN1-AS1 may be a novel biomarker and therapeutic target for NSCLC patients.
Keywords: DEPDC1; NSCLC; lncRNA KTN1-AS1; metastasis; miR-23b.
Conflict of interest statement
Figures



References
-
- Zhang Y, Zhang M, Jiang Y, Li X, He Y, Zeng P, Guo Z, Chang Y, Luo H, Liu Y, Hao C, Wang H, Zhang G, Zhang L. Lentinan as an immunotherapeutic for treating lung cancer: a review of 12 years clinical studies in China. J Cancer Res Clin Oncol. 2018; 144:2177–86. 10.1007/s00432-018-2718-1 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

