Development and Evaluation of a Single Dye Duplex Droplet Digital PCR Assay for the Rapid Detection and Quantification of Mycobacterium tuberculosis
- PMID: 32397601
- PMCID: PMC7284639
- DOI: 10.3390/microorganisms8050701
Development and Evaluation of a Single Dye Duplex Droplet Digital PCR Assay for the Rapid Detection and Quantification of Mycobacterium tuberculosis
Abstract
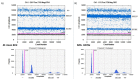
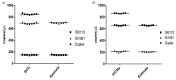
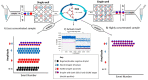
Droplet digital PCR (ddPCR) is a third generation of PCR that was recently developed to overcome the challenges of real-time fluorescence-based quantitative PCR (qPCR) in absolute quantification of pathogens. Few studies have been done on tuberculosis (TB) detection and quantification using ddPCR despite its many advantages over qPCR. From the few studies, none explores a single dye duplex assay for the detection and quantification of TB. In this study, steps toward developing and evaluating a duplex single dye (FAM) assay for detecting two targets (IS6110 and IS1081) are clearly described using simplex and duplex experiments. To achieve this, various parameters are investigated, including annealing temperature, primer and probe concentration, sensitivity and specificity, sample concentration, and inter/intra-assay variability. From the results, primer and probe concentration, annealing temperature, and sample concentration have an effect on the position and separation of droplets in both simplex and duplex assays. The copies of target genes in a duplex assay can be estimated accurately using the threshold tool with little inter-assay (CV <1%) and intra-assay (CV <6%) variability when compared to simplex assays. The ddPCR assay specificity and sensitivity are both 100% when compared to qPCR. This work shows steps toward the detection and quantification of two targets in a single channel, enabling higher multiplexing to include more targets in future works.
Keywords: Detection; Duplex assay; IS1081; IS6110; Mycobacterium tuberculosis; Quantification; Simplex assay; ddPCR; qPCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- WHO Global Tuberculosis Report. [(accessed on 22 March 2019)];2018 Available online: https://www.who.int/tb/publications/global_report/en/
Grants and funding
LinkOut - more resources
Full Text Sources

