Hypermethylation and decreased expression of TMEM240 are potential early-onset biomarkers for colorectal cancer detection, poor prognosis, and early recurrence prediction
- PMID: 32398064
- PMCID: PMC7218647
- DOI: 10.1186/s13148-020-00855-z
Hypermethylation and decreased expression of TMEM240 are potential early-onset biomarkers for colorectal cancer detection, poor prognosis, and early recurrence prediction
Abstract
Background: Gene silencing by aberrant DNA methylation of promoter regions remains the most dominant phenomenon occurring during tumorigenesis. Improving the early diagnosis, prognosis, and recurrence prediction of colorectal cancer using noninvasive aberrant DNA methylation biomarkers has encouraging potential. The aim of this study is to characterize the DNA methylation of the promoter region of TMEM240, as well as gene expression and its effect on cell biological functions and its applications in early detection and outcome prediction.
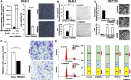
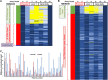
Results: Highly methylated CpG sites were identified in the TMEM240 gene by Illumina methylation 450K arrays in 26 Taiwanese patient paired samples and 38 paired samples from The Cancer Genome Atlas (TCGA) colorectal cancer dataset. Transient transfection and knockdown of TMEM240 were performed to demonstrate the role of TMEM240 in colorectal cancer cells. The data showed that TMEM240 could lead to G1 cell cycle arrest, repress cancer cell proliferation, and inhibit cancer cell migration. The quantitative methylation-specific real-time polymerase chain reaction (PCR) results revealed that 87.8% (480 of 547) of the colorectal cancer tumors had hypermethylated TMEM240, and this was also found in benign tubular adenomas (55.6%). Circulating cell-free methylated TMEM240 was detected in 13 of 25 (52.0%) Taiwanese colorectal cancer patients but in fewer (28.6%) healthy controls. In 72.0% (85/118) of tissue samples, TMEM240 mRNA expression was lower in Taiwanese CRC tumor tissues than in normal colorectal tissues according to real-time reverse transcription PCR results, and this was also found in benign tubular adenomas (44.4%). The TMEM240 protein was analyzed in South Korean and Chinese CRC patient samples using immunohistochemistry. The results exhibited low protein expression in 91.7% (100/109) of tumors and 75.0% (24/32) of metastatic tumors but exhibited high expression in 75.0% (6/8) of normal colon tissues. Multivariate Cox proportional hazards regression analysis found that mRNA expression of TMEM240 was significantly associated with overall, cancer-specific, and recurrence-free survival (p = 0.012, 0.007, and 0.022, respectively).
Conclusions: Alterations in TMEM240 are commonly found in Western and Asian populations and can potentially be used for early prediction and as poor prognosis and early-recurrence biomarkers in colorectal cancer.
Keywords: Circulating cell-free DNA; DNA methylation; Early detection; Early onset; Prognostic marker; TMEM240; ccfDNA; cmDNA.
Conflict of interest statement
There are no conflicts of interest.
Figures





References
-
- Taiwan Ministry of Health and Welfare. 2018 Taiwan health and welfare report. 2019.
-
- Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 13/12/2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

