The potential role of RNA N6-methyladenosine in Cancer progression
- PMID: 32398132
- PMCID: PMC7216508
- DOI: 10.1186/s12943-020-01204-7
The potential role of RNA N6-methyladenosine in Cancer progression
Abstract
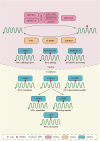
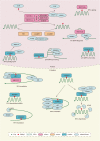
N6-methyladenosine (m6A) is considered the most common, abundant, and conserved internal transcript modification, especially in eukaryotic messenger RNA (mRNA). m6A is installed by m6A methyltransferases (METTL3/14, WTAP, RBM15/15B, VIRMA and ZC3H13, termed "writers"), removed by demethylases (FTO, ALKBH5, and ALKBH3, termed "erasers"), and recognized by m6A-binding proteins (YTHDC1/2, YTHDF1/2/3, IGF2BP1/2/3, HNRNP, and eIF3, termed "readers"). Accumulating evidence suggests that m6A RNA methylation greatly impacts RNA metabolism and is involved in the pathogenesis of many kinds of diseases, including cancers. In this review, we focus on the physiological functions of m6A modification and its related regulators, as well as on the potential biological roles of these elements in human tumors.
Keywords: Cancer progression; Molecular mechanisms; N6-methyladenosine (m6A).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

