Structural optimization of a TNFR1-selective antagonistic TNFα mutant to create new-modality TNF-regulating biologics
- PMID: 32398258
- PMCID: PMC7363114
- DOI: 10.1074/jbc.RA120.012723
Structural optimization of a TNFR1-selective antagonistic TNFα mutant to create new-modality TNF-regulating biologics
Abstract
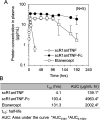
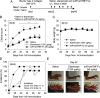
Excessive activation of the proinflammatory cytokine tumor necrosis factor-α (TNFα) is a major cause of autoimmune diseases, including rheumatoid arthritis. TNFα induces immune responses via TNF receptor 1 (TNFR1) and TNFR2. Signaling via TNFR1 induces proinflammatory responses, whereas TNFR2 signaling is suggested to suppress the pathophysiology of inflammatory diseases. Therefore, selective inhibition of TNFR1 signaling and preservation of TNFR2 signaling activities may be beneficial for managing autoimmune diseases. To this end, we developed a TNFR1-selective, antagonistic TNFα mutant (R1antTNF). Here, we developed an R1antTNF derivative, scR1antTNF-Fc, which represents a single-chain form of trimeric R1antTNF with a human IgG-Fc domain. scR1antTNF-Fc had properties similar to those of R1antTNF, including TNFR1-selective binding avidity, TNFR1 antagonistic activity, and thermal stability, and had a significantly extended plasma t1/2in vivo In a murine rheumatoid arthritis model, scR1antTNF-Fc and 40-kDa PEG-scR1antTNF (a previously reported PEGylated form) delayed the onset of collagen-induced arthritis, suppressed arthritis progression in mice, and required a reduced frequency of administration. Interestingly, with these biologic treatments, we observed an increased ratio of regulatory T cells to conventional T cells in lymph nodes compared with etanercept, a commonly used TNF inhibitor. Therefore, scR1antTNF-Fc and 40-kDa PEG-scR1antTNF indirectly induced immunosuppression. These results suggest that selective TNFR1 inhibition benefits the management of autoimmune diseases and that R1antTNF derivatives hold promise as new-modality TNF-regulating biologics.
Keywords: antagonist; arthritis; autoimmune disease; cytokine; drug delivery; drug design; forkhead box P3 (FOXP3); inflammation; inhibition mechanism; protein engineering; single-chain; tumor necrosis factor (TNF).
© 2020 Inoue et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
-
- Gottenberg J. E., Brocq O., Perdriger A., Lassoued S., Berthelot J. M., Wendling D., Euller-Ziegler L., Soubrier M., Richez C., Fautrel B., Constantin A. L., Mariette X., Morel J., Gilson M., Cormier G., et al. (2016) Non-TNF-targeted biologic vs a second anti-TNF drug to treat rheumatoid arthritis in patients with insufficient response to a first anti-TNF drug: A randomized clinical trial. JAMA 316, 1172–1180 10.1001/jama.2016.13512 - DOI - PubMed
-
- van der Bijl A. E., Breedveld F. C., Antoni C. E., Kalden J. R., Kary S., Burmester G. R., Beckmann C., Unnebrink K., and Kupper H. (2008) An open-label pilot study of the effectiveness of adalimumab in patients with rheumatoid arthritis and previous infliximab treatment: Relationship to reasons for failure and anti-infliximab antibody status. Clin. Rheumatol. 27, 1021–1028 10.1007/s10067-008-0866-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

