Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma
- PMID: 32398863
- PMCID: PMC7608103
- DOI: 10.1038/s41422-020-0333-6
Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma
Erratum in
-
Author Correction: Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma.Cell Res. 2022 Apr;32(4):415-416. doi: 10.1038/s41422-022-00625-x. Cell Res. 2022. PMID: 35177820 Free PMC article. No abstract available.
Abstract
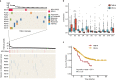
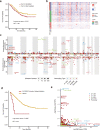
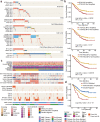
Esophageal squamous cell carcinoma (ESCC) is a poor-prognosis cancer type with limited understanding of its molecular etiology. Using 508 ESCC genomes, we identified five novel significantly mutated genes and uncovered mutational signature clusters associated with metastasis and patients' outcomes. Several functional assays implicated that NFE2L2 may act as a tumor suppressor in ESCC and that mutations in NFE2L2 probably impaired its tumor-suppressive function, or even conferred oncogenic activities. Additionally, we found that the NFE2L2 mutations were significantly associated with worse prognosis of ESCC. We also identified potential noncoding driver mutations including hotspot mutations in the promoter region of SLC35E2 that were correlated with worse survival. Approximately 5.9% and 15.2% of patients had high tumor mutation burden or actionable mutations, respectively, and may benefit from immunotherapy or targeted therapies. We found clinically relevant coding and noncoding genomic alterations and revealed three major subtypes that robustly predicted patients' outcomes. Collectively, we report the largest dataset of genomic profiling of ESCC useful for developing ESCC-specific biomarkers for diagnosis and treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Chen W, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. - PubMed
-
- Lin Y, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann. Epidemiol. 2017;27:215–221. - PubMed
-
- Gao YB, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:1097–1102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

