State-of-the-art tools to identify druggable protein ligand of SARS-CoV-2
- PMID: 32399095
- PMCID: PMC7212236
- DOI: 10.5114/aoms.2020.94046
State-of-the-art tools to identify druggable protein ligand of SARS-CoV-2
Abstract
Introduction: The SARS-CoV-2 (previously 2019-nCoV) outbreak in Wuhan, China and other parts of the world affects people and spreads coronavirus disease 2019 (COVID-19) through human-to-human contact, with a mortality rate of > 2%. There are no approved drugs or vaccines yet available against SARS-CoV-2.
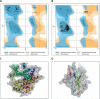
Material and methods: State-of-the-art tools based on in-silico methods are a cost-effective initial approach for identifying appropriate ligands against SARS-CoV-2. The present study developed the 3D structure of the envelope and nucleocapsid phosphoprotein of SARS-CoV-2, and molecular docking analysis was done against various ligands.
Results: The highest log octanol/water partition coefficient, high number of hydrogen bond donors and acceptors, lowest non-bonded interaction energy between the receptor and the ligand, and high binding affinity were considered for the best ligand for the envelope (mycophenolic acid: log P = 3.00; DG = -10.2567 kcal/mol; pKi = 7.713 µM) and nucleocapsid phosphoprotein (1-[(2,4-dichlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid: log P = 2.901; DG = -12.2112 kcal/mol; pKi = 7.885 µM) of SARS-CoV-2.
Conclusions: The study identifies the most potent compounds against the SARS-CoV-2 envelope and nucleocapsid phosphoprotein through state-of-the-art tools based on an in-silico approach. A combination of these two ligands could be the best option to consider for further detailed studies to develop a drug for treating patients infected with SARS-CoV-2, COVID-19.
Keywords: COVID-19; SARS-CoV-2; druggable protein; envelope protein; ligand; molecular docking; nucleocapsid phosphoprotein; phylogenetic tree.
Copyright: © 2020 Termedia & Banach.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO, 2020a Naming the coronavirus disease (COVID-19) and the virus that causes it, Accessed 23 Mar 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica....
-
- Cárdenas-Conejo Y, Liñan-Rico A, García-Rodríguez DA, Centeno-Leija S, Serrano-Posada H. An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human-pathogenic coronavirus (SARS-CoV2) J Med Virol. 2020 doi: 10.1002/jmv.25758. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
