State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2
- PMID: 32399096
- PMCID: PMC7212215
- DOI: 10.5114/aoms.2020.94567
State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2
Abstract
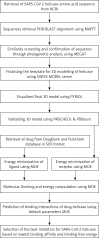
Introduction: The extreme health and economic problems in the world due to the SARS-CoV-2 infection have led to an urgent need to identify potential drug targets for treating coronavirus disease 2019 (COVID-19). The present state-of-the-art tool-based screening was targeted to identify drug targets among clinically approved drugs by uncovering SARS-CoV-2 helicase inhibitors through molecular docking analysis.
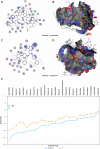
Material and methods: Helicase is a vital viral replication enzyme, which unwinds nucleic acids and separates the double-stranded nucleic acids into single-stranded nucleic acids. Hence, the SARS-CoV-2 helicase protein 3D structure was predicted, validated, and used to screen the druggable targets among clinically approved drugs such as protease inhibitor, nucleoside reverse transcriptase inhibitor, and non-nucleoside reverse transcriptase inhibitors, used to treat HIV infection using molecular docking analysis.
Results: Interaction with SARS-CoV-2 helicase, approved drugs, vapreotide (affinity: -12.88; S score: -9.84 kcal/mol), and atazanavir (affinity: -11.28; S score: -9.32 kcal/mol), approved drugs for treating AIDS-related diarrhoea and HIV infection, respectively, are observed with significantly low binding affinity and MOE score or binding free energy. The functional binding pockets of the clinically approved drugs on SARS-CoV-2 helicase protein molecule suggest that vapreotide and atazanavir may interrupt the activities of the SARS-CoV-2 helicase.
Conclusions: The study suggests that vapreotide may be a choice of drug for wet lab studies to inhibit the infection of SARS-CoV-2.
Keywords: COVID-19; SARS-CoV-2; antiretroviral agents; clinically approved drugs; helicase; molecular docking.
Copyright: © 2020 Termedia & Banach.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- WHO 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019; Accessed on 08 Apr 2020.
-
- World Health OrganizationWHO . World Health Organization; 2019. Clinical management of severe acute respiratory infection when Middle East respiratory syndrome coronavirus (MERS-CoV) infection is suspected: interim guidance (No. WHO/MERS/Clinical/15.1 Revision 1)
LinkOut - more resources
Full Text Sources
Miscellaneous
