A role for MIR828 in pineapple fruit development
- PMID: 32399197
- PMCID: PMC7194464
- DOI: 10.12688/f1000research.21779.2
A role for MIR828 in pineapple fruit development
Abstract
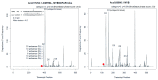
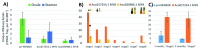
Chen et al. ( Nature Genet. 51: 1549-1558; Oct. 2019) sequenced Ananas comosus var. bracteatus accession CB5, cultivated for its bright pink-to-red colored fruit, and yellow-fleshed A. comosus accession F153, reporting an improved F153 reference assembly while annotating MICRORNA (MIRNA) loci and gene family expressions relevant to lignin and anthocyanin biosynthesis. An independent article (Xiong et al.Sci. Rep. 8: 1947; 2018) reported var. bracteatus MIRNAs but not MIR828, a negative regulator of anthocyanin and polyphenolics biosynthesis by targeting MYB transcription factors associated with UV light- and sugar-signaling in dicots. MIR828 has been reported in gymnosperms, Amborella (sister to flowering plants), and basal monocot orders Liliales, Asparagales, Zingiberales, Arecales, but not in the Poales, a sister order comprising grasses and ~3,000 species of bromeliads including pineapple. Here I show MIR828 exists in pineapple and directs post-transcriptional gene silencing of mRNAs encoding MYB family members with inferred function to regulate the conspicuous red fruit trait in var. bracteatus. MIR828 plesiomorphy (an ancient basal trait) may shed light on monocot apomorphic fruit development, postulated for 21 monocot families with fleshy fruits as due to homoplasy/convergence driven by tropical climate and/or enticements to vertebrate endozoic seed dispersers.
Keywords: RNA interference; anthocyanins; evolution; fruit development; microRNAs.
Copyright: © 2020 Rock CD.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
-
- Zheng Y, Li T, Xu Z, et al. : Identification of microRNAs, phasiRNAs and their targets in pineapple. Trop Plant Biol. 2016;9:176–186. 10.1007/s12042-016-9173-4 - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

