Prp5-Spt8/Spt3 interaction mediates a reciprocal coupling between splicing and transcription
- PMID: 32399566
- PMCID: PMC7293005
- DOI: 10.1093/nar/gkaa311
Prp5-Spt8/Spt3 interaction mediates a reciprocal coupling between splicing and transcription
Abstract
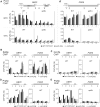
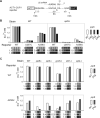
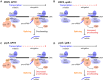
Transcription and pre-mRNA splicing are coupled to promote gene expression and regulation. However, mechanisms by which transcription and splicing influence each other are still under investigation. The ATPase Prp5p is required for pre-spliceosome assembly and splicing proofreading at the branch-point region. From an open UV mutagenesis screen for genetic suppressors of prp5 defects and subsequent targeted testing, we identify components of the TBP-binding module of the Spt-Ada-Gcn5 Acetyltransferase (SAGA) complex, Spt8p and Spt3p. Spt8Δ and spt3Δ rescue the cold-sensitivity of prp5-GAR allele, and prp5 mutants restore growth of spt8Δ and spt3Δ strains on 6-azauracil. By chromatin immunoprecipitation (ChIP), we find that prp5 alleles decrease recruitment of RNA polymerase II (Pol II) to an intron-containing gene, which is rescued by spt8Δ. Further ChIP-seq reveals that global effects on Pol II-binding are mutually rescued by prp5-GAR and spt8Δ. Inhibited splicing caused by prp5-GAR is also restored by spt8Δ. In vitro assays indicate that Prp5p directly interacts with Spt8p, but not Spt3p. We demonstrate that Prp5p's splicing proofreading is modulated by Spt8p and Spt3p. Therefore, this study reveals that interactions between the TBP-binding module of SAGA and the spliceosomal ATPase Prp5p mediate a balance between transcription initiation/elongation and pre-spliceosome assembly.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Maniatis T., Reed R.. An extensive network of coupling among gene expression machines. Nature. 2002; 416:499–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

