Multiple weak interactions between BvgA~P and ptx promoter DNA strongly activate transcription of pertussis toxin genes in Bordetella pertussis
- PMID: 32401811
- PMCID: PMC7250471
- DOI: 10.1371/journal.ppat.1008500
Multiple weak interactions between BvgA~P and ptx promoter DNA strongly activate transcription of pertussis toxin genes in Bordetella pertussis
Abstract
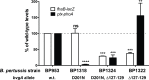
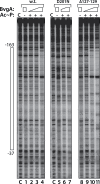
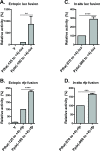
Pertussis toxin is the preeminent virulence factor and major protective antigen produced by Bordetella pertussis, the human respiratory pathogen and etiologic agent of whooping cough. Genes for its synthesis and export are encoded by the 12 kb ptx-ptl operon, which is under the control of the pertussis promoter, Pptx. Expression of this operon, like that of all other known protein virulence factors, is regulated by the BvgAS two-component global regulatory system. Although Pptx has been studied for years, characterization of its promoter architecture vis-à-vis BvgA-binding has lagged behind that of other promoters, mainly due to its lower affinity for BvgA~P. Here we take advantage of a mutant BvgA protein (Δ127-129), which enhances ptx transcription in B. pertussis and also demonstrates enhanced binding affinity to Pptx. By using this mutant protein labeled with FeBABE, binding of six head-to-head dimers of BvgA~P was observed, with a spacing of 22 bp, revealing a binding geometry similar to that of other BvgA-activated promoters carrying at least one strong binding site. All of these six BvgA-binding sites lack sequence features associated with strong binding. A genetic analysis indicated the degree to which each contributes to Pptx activity. Thus the weak/medium binding affinity of Pptx revealed in this study explains its lower responsiveness to phosphorylated BvgA, relative to other promoters containing a high affinity binding site, such as that of the fha operon.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter.J Bacteriol. 1997 Mar;179(5):1755-63. doi: 10.1128/jb.179.5.1755-1763.1997. J Bacteriol. 1997. PMID: 9045838 Free PMC article.
-
Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis.J Bacteriol. 1995 Nov;177(22):6486-91. doi: 10.1128/jb.177.22.6486-6491.1995. J Bacteriol. 1995. PMID: 7592424 Free PMC article.
-
The BvgAS Regulon of Bordetella pertussis.mBio. 2017 Oct 10;8(5):e01526-17. doi: 10.1128/mBio.01526-17. mBio. 2017. PMID: 29018122 Free PMC article.
-
The Bordetella pertussis model of exquisite gene control by the global transcription factor BvgA.Microbiology (Reading). 2012 Jul;158(Pt 7):1665-1676. doi: 10.1099/mic.0.058941-0. Epub 2012 May 24. Microbiology (Reading). 2012. PMID: 22628479 Free PMC article. Review.
-
Signal transduction and virulence regulation in Bordetella pertussis.Microbiologia. 1996 Jun;12(2):185-96. Microbiologia. 1996. PMID: 8767703 Review.
Cited by
-
AB5 Enterotoxin-Mediated Pathogenesis: Perspectives Gleaned from Shiga Toxins.Toxins (Basel). 2022 Jan 16;14(1):62. doi: 10.3390/toxins14010062. Toxins (Basel). 2022. PMID: 35051039 Free PMC article. Review.
-
Four single-basepair mutations in the ptx promoter of Bordetella bronchiseptica are sufficient to activate the expression of pertussis toxin.Sci Rep. 2021 Apr 30;11(1):9373. doi: 10.1038/s41598-021-88852-x. Sci Rep. 2021. PMID: 33931696 Free PMC article.
-
Conformational change of the Bordetella response regulator BvgA accompanies its activation of the B. pertussis virulence gene fhaB.Comput Struct Biotechnol J. 2022 Nov 6;20:6431-6442. doi: 10.1016/j.csbj.2022.10.042. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467586 Free PMC article.
-
Lonidamine, a Novel Modulator for the BvgAS System of Bordetella Species.Microbiol Immunol. 2025 Mar;69(3):133-147. doi: 10.1111/1348-0421.13193. Epub 2024 Dec 15. Microbiol Immunol. 2025. PMID: 39674913 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

