KAP1 Is a Chromatin Reader that Couples Steps of RNA Polymerase II Transcription to Sustain Oncogenic Programs
- PMID: 32402252
- PMCID: PMC7305985
- DOI: 10.1016/j.molcel.2020.04.024
KAP1 Is a Chromatin Reader that Couples Steps of RNA Polymerase II Transcription to Sustain Oncogenic Programs
Abstract
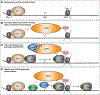
Precise control of the RNA polymerase II (RNA Pol II) cycle, including pausing and pause release, maintains transcriptional homeostasis and organismal functions. Despite previous work to understand individual transcription steps, we reveal a mechanism that integrates RNA Pol II cycle transitions. Surprisingly, KAP1/TRIM28 uses a previously uncharacterized chromatin reader cassette to bind hypo-acetylated histone 4 tails at promoters, guaranteeing continuous progression of RNA Pol II entry to and exit from the pause state. Upon chromatin docking, KAP1 first associates with RNA Pol II and then recruits a pathway-specific transcription factor (SMAD2) in response to cognate ligands, enabling gene-selective CDK9-dependent pause release. This coupling mechanism is exploited by tumor cells to aberrantly sustain transcriptional programs commonly dysregulated in cancer patients. The discovery of a factor integrating transcription steps expands the functional repertoire by which chromatin readers operate and provides mechanistic understanding of transcription regulation, offering alternative therapeutic opportunities to target transcriptional dysregulation.
Keywords: CDK9; KAP1; RNA polymerase II; SMAD; TGF-β; TRIM28; cancer; chromatin reader; epigenetics; pausing.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures






References
-
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, and Hill CS (2011). Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell 43, 85–96. - PubMed
-
- Ahn SH, Kim M, and Buratowski S (2004). Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3’ end processing. Mol Cell 13, 67–76. - PubMed
-
- Allis CD, and Jenuwein T (2016). The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

