Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation
- PMID: 32402267
- PMCID: PMC7217133
- DOI: 10.1016/j.cmet.2020.04.015
Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation
Abstract
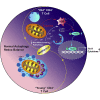
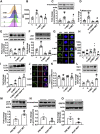
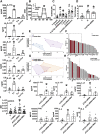
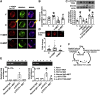
Age is a non-modifiable risk factor for the inflammation that underlies age-associated diseases; thus, anti-inflammaging drugs hold promise for increasing health span. Cytokine profiling and bioinformatic analyses showed that Th17 cytokine production differentiates CD4+ T cells from lean, normoglycemic older and younger subjects, and mimics a diabetes-associated Th17 profile. T cells from older compared to younger subjects also had defects in autophagy and mitochondrial bioenergetics that associate with redox imbalance. Metformin ameliorated the Th17 inflammaging profile by increasing autophagy and improving mitochondrial bioenergetics. By contrast, autophagy-targeting siRNA disrupted redox balance in T cells from young subjects and activated the Th17 profile by activating the Th17 master regulator, STAT3, which in turn bound IL-17A and F promoters. Mitophagy-targeting siRNA failed to activate the Th17 profile. We conclude that metformin improves autophagy and mitochondrial function largely in parallel to ameliorate a newly defined inflammaging profile that echoes inflammation in diabetes.
Keywords: T cells; autophagy; inflammaging; metformin; mitochondria.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



Comment in
-
CD4+ T Cells at the Center of Inflammaging.Cell Metab. 2020 Jul 7;32(1):4-5. doi: 10.1016/j.cmet.2020.04.016. Epub 2020 May 12. Cell Metab. 2020. PMID: 32402265 Free PMC article.
-
Next steps in mechanisms of inflammaging.Autophagy. 2020 Dec;16(12):2285-2286. doi: 10.1080/15548627.2020.1822089. Epub 2020 Sep 22. Autophagy. 2020. PMID: 32960694 Free PMC article.
References
-
- Almeida L., Lochner M., Berod L., Sparwasser T. Metabolic pathways in T cell activation and lineage differentiation. Semin. Immunol. 2016;28:514–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

