A RT-PCR assay for the detection of coronaviruses from four genera
- PMID: 32403008
- PMCID: PMC7192118
- DOI: 10.1016/j.jcv.2020.104391
A RT-PCR assay for the detection of coronaviruses from four genera
Abstract
Background: During the past two decades, three novel coronaviruses (CoVs) have emerged to cause international human epidemics with severe morbidity. CoVs have also emerged to cause severe epidemics in animals. A better understanding of the natural hosts and genetic diversity of CoVs are needed to help mitigate these threats.
Objective: To design and evaluate a molecular diagnostic tool for detection and identification of all currently recognized and potentially future emergent CoVs from the Orthocoronavirinae subfamily.
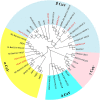
Study design and results: We designed a semi-nested, reverse transcription RT-PCR assay based upon 38 published genome sequences of human and animal CoVs. We evaluated this assay with 14 human and animal CoVs and 11 other non-CoV respiratory viruses. Through sequencing the assay's target amplicon, the assay correctly identified each of the CoVs; no cross-reactivity with 11 common respiratory viruses was observed. The limits of detection ranged from 4 to 4 × 102 copies/reaction, depending on the CoV species tested. To assess the assay's clinical performance, we tested a large panel of previously studied specimens: 192 human respiratory specimens from pneumonia patients, 5 clinical specimens from COVID-19 patients, 81 poultry oral secretion specimens, 109 pig slurry specimens, and 31 aerosol samples from a live bird market. The amplicons of all RT-PCR-positive samples were confirmed by Sanger sequencing. Our assay performed well with all tested specimens across all sample types.
Conclusions: This assay can be used for detection and identification of all previously recognized CoVs, including SARS-CoV-2, and potentially any emergent CoVs in the Orthocoronavirinae subfamily.
Keywords: COVID-19; Coronavirus; Emerging; Infectious diseases; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors have declared that no competing financial interests exist.
Figures


References
-
- ICTV: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-... (Accessed on April 9, 2020).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

