The Microbiome in Cystic Fibrosis Pulmonary Disease
- PMID: 32403302
- PMCID: PMC7288443
- DOI: 10.3390/genes11050536
The Microbiome in Cystic Fibrosis Pulmonary Disease
Abstract
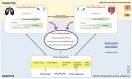
Cystic fibrosis (CF) is a genetic disease with mutational changes leading to profound dysbiosis, both pulmonary and intestinal, from a very young age. This dysbiosis plays an important role in clinical manifestations, particularly in the lungs, affected by chronic infection. The range of microbiological tools has recently been enriched by metagenomics based on next-generation sequencing (NGS). Currently applied essentially in a gene-targeted manner, metagenomics has enabled very exhaustive description of bacterial communities in the CF lung niche and, to a lesser extent, the fungi. Aided by progress in bioinformatics, this now makes it possible to envisage shotgun sequencing and opens the door to other areas of the microbial world, the virome, and the archaeome, for which almost everything remains to be described in cystic fibrosis. Paradoxically, applying NGS in microbiology has seen a rebirth of bacterial culture, but in an extended manner (culturomics), which has proved to be a perfectly complementary approach to NGS. Animal models have also proved indispensable for validating microbiome pathophysiological hypotheses. Description of pathological microbiomes and correlation with clinical status and therapeutics (antibiotic therapy, cystic fibrosis transmembrane conductance regulator (CFTR) modulators) revealed the richness of microbiome data, enabling description of predictive and follow-up biomarkers. Although monogenic, CF is a multifactorial disease, and both genotype and microbiome profiles are crucial interconnected factors in disease progression. Microbiome-genome interactions are thus important to decipher.
Keywords: cystic fibrosis; gut–lung axis; lung microbiome; metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

