Diverse LEF/TCF Expression in Human Colorectal Cancer Correlates with Altered Wnt-Regulated Transcriptome in a Meta-Analysis of Patient Biopsies
- PMID: 32403323
- PMCID: PMC7288467
- DOI: 10.3390/genes11050538
Diverse LEF/TCF Expression in Human Colorectal Cancer Correlates with Altered Wnt-Regulated Transcriptome in a Meta-Analysis of Patient Biopsies
Abstract
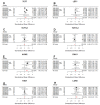
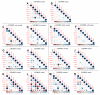
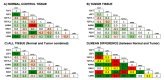
Aberrantly activated Wnt signaling causes cellular transformation that can lead to human colorectal cancer. Wnt signaling is mediated by Lymphoid Enhancer Factor/T-Cell Factor (LEF/TCF) DNA-binding factors. Here we investigate whether altered LEF/TCF expression is conserved in human colorectal tumor sample and may potentially be correlated with indicators of cancer progression. We carried out a meta-analysis of carefully selected publicly available gene expression data sets with paired tumor biopsy and adjacent matched normal tissues from colorectal cancer patients. Our meta-analysis confirms that among the four human LEF/TCF genes, LEF1 and TCF7 are preferentially expressed in tumor biopsies, while TCF7L2 and TCF7L1 in normal control tissue. We also confirm positive correlation of LEF1 and TCF7 expression with hallmarks of active Wnt signaling (i.e., AXIN2 and LGR5). We are able to correlate differential LEF/TCF gene expression with distinct transcriptomes associated with cell adhesion, extracellular matrix organization, and Wnt receptor feedback regulation. We demonstrate here in human colorectal tumor sample correlation of altered LEF/TCF gene expression with quantitatively and qualitatively different transcriptomes, suggesting LEF/TCF-specific transcriptional regulation of Wnt target genes relevant for cancer progression and survival. This bioinformatics analysis provides a foundation for future more detailed, functional, and molecular analyses aimed at dissecting such functional differences.
Keywords: LEF; TCF; Wnt; colorectal cancer; transcriptome.
Conflict of interest statement
The authors are not aware of any conflict of interest.
Figures
Similar articles
-
Transcriptional activity mediated by β-CATENIN and TCF/LEF family members is completely dispensable for survival and propagation of multiple human colorectal cancer cell lines.Sci Rep. 2023 Jan 6;13(1):287. doi: 10.1038/s41598-022-27261-0. Sci Rep. 2023. PMID: 36609428 Free PMC article.
-
Lymphoid enhancer factor/T cell factor expression in colorectal cancer.Cancer Metastasis Rev. 2004 Jan-Jun;23(1-2):41-52. doi: 10.1023/a:1025858928620. Cancer Metastasis Rev. 2004. PMID: 15000148 Review.
-
Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development.Development. 2005 Dec;132(24):5375-85. doi: 10.1242/dev.02152. Epub 2005 Nov 16. Development. 2005. PMID: 16291789
-
TCF7L1 regulates colorectal cancer cell migration by repressing GAS1 expression.Sci Rep. 2024 May 30;14(1):12477. doi: 10.1038/s41598-024-63346-8. Sci Rep. 2024. PMID: 38816533 Free PMC article.
-
Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials.Crit Rev Eukaryot Gene Expr. 2011;21(3):207-36. doi: 10.1615/critreveukargeneexpr.v21.i3.10. Crit Rev Eukaryot Gene Expr. 2011. PMID: 22111711 Free PMC article. Review.
Cited by
-
Angiogenesis-Related Functions of Wnt Signaling in Colorectal Carcinogenesis.Cancers (Basel). 2020 Dec 2;12(12):3601. doi: 10.3390/cancers12123601. Cancers (Basel). 2020. PMID: 33276489 Free PMC article. Review.
-
Functional analysis of CTLA4 promoter variant and its possible implication in colorectal cancer immunotherapy.Front Med (Lausanne). 2023 Aug 3;10:1160368. doi: 10.3389/fmed.2023.1160368. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37601778 Free PMC article.
-
The WNT/β-catenin dependent transcription: A tissue-specific business.WIREs Mech Dis. 2021 May;13(3):e1511. doi: 10.1002/wsbm.1511. Epub 2020 Oct 21. WIREs Mech Dis. 2021. PMID: 33085215 Free PMC article. Review.
-
Resveratrol inhibits development of colorectal adenoma via suppression of LEF1; comprehensive analysis with connectivity map.Cancer Sci. 2022 Dec;113(12):4374-4384. doi: 10.1111/cas.15576. Epub 2022 Sep 20. Cancer Sci. 2022. PMID: 36082704 Free PMC article.
-
TCF7L1 Regulates LGR5 Expression in Colorectal Cancer Cells.Genes (Basel). 2023 Feb 14;14(2):481. doi: 10.3390/genes14020481. Genes (Basel). 2023. PMID: 36833408 Free PMC article.
References
-
- Hoppler S., Moon R.T. Wnt Signaling in Development and Disease: Molecular Mechanisms and Biological Functions. Wiley Blackwell; Hoboken, NJ, USA: 2014. p. 459.
-
- Hoppler S., Nakamura Y. Cell-to-Cell Signalling in Development: Wnt Signalling. eLS. 2014 doi: 10.1002/9780470015902.a0002331.pub2. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

